
Page 161
Notes:
conferenceseries
.com
Volume 10, Issue 8 (Suppl)
J Proteomics Bioinform, an open access journal
ISSN: 0974-276X
Structural Biology 2017
September 18-20, 2017
9
th
International Conference on
Structural Biology
September 18-20, 2017 Zurich, Switzerland
Pradeep Pant et al., J Proteomics Bioinform 2017, 10:8(Suppl)
DOI: 10.4172/0974-276X-C1-0101
Design and characterization of symmetric nucleic acids
via
molecular dynamics simulations
Pradeep Pant
and
B Jayaram
Indian Institute of Technology Delhi, India
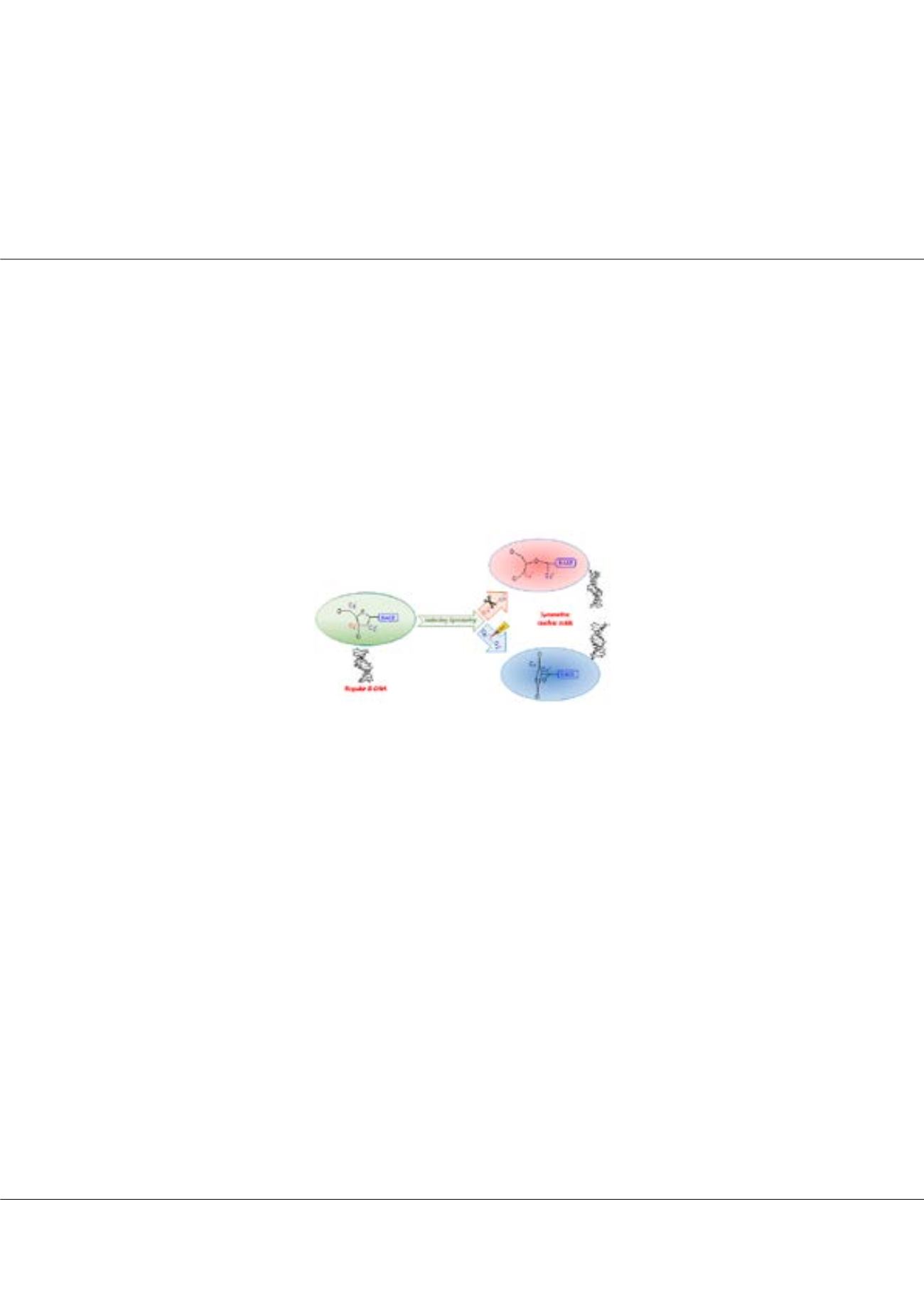
D
irectionality (5’→3’) is so fundamental to the nucleic acid architecture and is essential for replication and transcription. We
observed that this asymmetry can be manipulated either by breaking (C3’ to C2’) or making (C5’ to C2’) chemical bond
in each nucleotide unit leading to symmetric nucleic acids. Keeping their potential synthetic and therapeutic interest in mind,
we designed a few novel symmetric nucleic acids. We investigated their conformational stability and flexibility
via
detailed all
atom explicit solvent 100-ns long molecular dynamics simulations and compared the resulting structures with that of regular
B-DNA. Quite interestingly, some of the symmetric nucleic acids retain the overall double helical structure indicating their
potential for integration in physiological DNA without causing major structural perturbations.
Biography
Pradeep Pant has done his Master’s in Chemistry from Indian Institute of Technology Delhi. He joined in the Department of Chemistry as a PhD scholar under the
supervision of Prof. B. Jayaram in 2014. Currently, he is focusing on theoretical studies on the structure, dynamics and energetics of nucleic acid-protein/ligand
interactions.
pradeep.pant25@gmail.com















