
Page 103
Notes:
conferenceseries
.com
Volume 10, Issue 8 (Suppl)
J Proteomics Bioinform, an open access journal
ISSN: 0974-276X
Structural Biology 2017
September 18-20, 2017
9
th
International Conference on
Structural Biology
September 18-20, 2017 Zurich, Switzerland
Steven Hayward, J Proteomics Bioinform 2017, 10:8(Suppl)
DOI: 10.4172/0974-276X-C1-0100
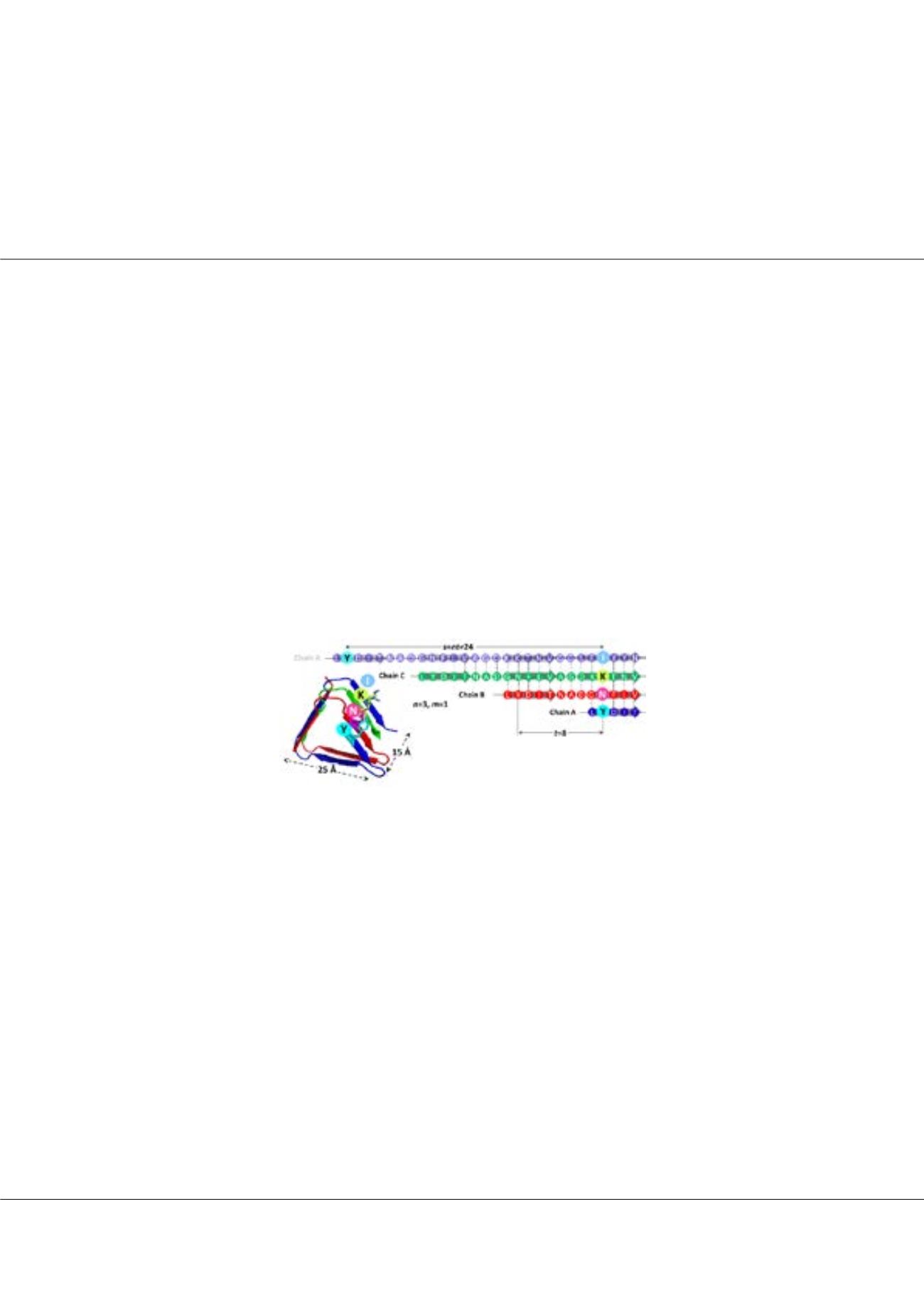
Geometrical principles of homeric β-barrels and β-helices: Applications to modelling amyloid
protofilaments
Steven Hayward
University of East Anglia, UK
E
xamples of homomeric β-helices and β-barrels have recently emerged. Here we have generalized the theory for the shear
number in β-barrels to encompass β-helices and homomeric structures. We introduce the concept of the β-strip which
comprises neighboring strands, parallel or antiparallel and forms the repeating unit that builds the helix. In this context,
the shear number is interpreted as the sum of register shifts between neighboring β-strips. This more general approach has
allowed us to derive relationships between the helical width, helical pitch, angle between strand direction and helical axis,
mass per length, register shift, and number of strands. The validity and unifying power of the method is demonstrated with
known structures including the T4 phage spike, cylindrin, and the HET-s (218-289) prion. The relationships have allowed us
to predict register shift and number of strands in transthyretin and Alzheimer β (40) amyloid protofilaments from reported
dimensions measured by X-ray fiber diffraction which we have used to construct models that comprise a single strip of in-
register β-strands folded into a β-strip helix. The results suggest that both stabilization of an individual β-strip helix as a
protofilament subunit and growth of the protofilament by the joining of subunits end-to-end, would involve the association of
the same pair of sequence segments at the same register shift.
Biography
Steven Hayward is a Reader in Computational Biology at the University of East Anglia. He uses computational methods, including bioinformatics techniques and
simulation, to understand protein structure, dynamics and function. He has focused particularly on protein dynamics and developed the popular DynDom method for
the analysis of domain movements in proteins. Recently he has worked on the fundamental structure of amyloid fibrils and works on the development of interactive
tools for protein visualization and docking using haptics.
steven.hayward@uea.ac.ukFigure1:
T4 phage spike


















