
Page 94
conferenceseries
.com
Volume 10, Issue 8 (Suppl)
J Proteomics Bioinform, an open access journal
ISSN: 0974-276X
Structural Biology 2017
September 18-20, 2017
9
th
International Conference on
Structural Biology
September 18-20, 2017 Zurich, Switzerland
How does domain motion contribute to transition-state stabilization? Combinatorial thermodynamic
cycle analysis of conformational coupling during tryptophan activation
Charles W Carter
University of North Carolina at Chapel Hill, USA
E
nzyme mechanisms, especially those that couple NTP hydrolysis to mechanical work and information, use sophisticated
dynamic networks to transduce active-site chemistry into domain motions that change binding affinities. We measured
and cross-validated the energetics of such networks in B.
stearothermophilus
Tryptophanyl-tRNA synthetase (TrpRS) using
both multi-mutant and modular thermodynamic cycles. Coordinated domain motions develop shear in a core packing motif
conserved in >125 different protein superfamilies. Multi-dimensional combinatorial mutagenesis showed that four side chains
from this “molecular switch” move coordinately with the active-site Mg2+ ion in the transition state for amino acid activation.
A modular thermodynamic cycle consisting of full-length TrpRS, Urzyme, and Urzyme plus each of the two domains deleted
in the Urzyme gives similar energetics. These complementary experiments establish that catalysis and specificity in full-length
TrpRS are both coupled by 5 kcal/mole to: (i) the core packing region where domain movement generates shear, and (ii) the
simultaneous motion of the two domains relative to the Urzyme. Theory shows that the minimum action path algorithm
estimates thermodynamically meaningful contributions of domain movement to kinetic rates. Correlations between those
parameters, the experimental rates, and structural variations induced in the combinatorial mutants confirm that these
estimates are realistic. These results validate our previous conclusion that catalysis by Mg2+ ion is coupled to the overall
domain motion. Computational free energy surfaces demonstrate that TrpRS catalytic domain motion itself is endergonic
but is driven thermodynamically by PPi release. Comparison of the impact of combinatorial mutagenesis on pre-steady state
and steady-state rates confirm that dynamic active-site pre-organization endows TrpRS with the elusive conditionality of NTP
utilization on domain motion.
Biography
Charles W Carter is an X-ray Crystallographer who studies the origin, evolution, and structural biology of aminoacyl-tRNA synthetases. His research group
introduced the use of urzyme-highly conserved structural cores that retain large fractions of the transition-state stabilization free energies of full length enzymes as
experimental models of ancestral enzymes.
carter@med.unc.teduCharles W Carter, J Proteomics Bioinform 2017, 10:8(Suppl)
DOI: 10.4172/0974-276X-C1-0100
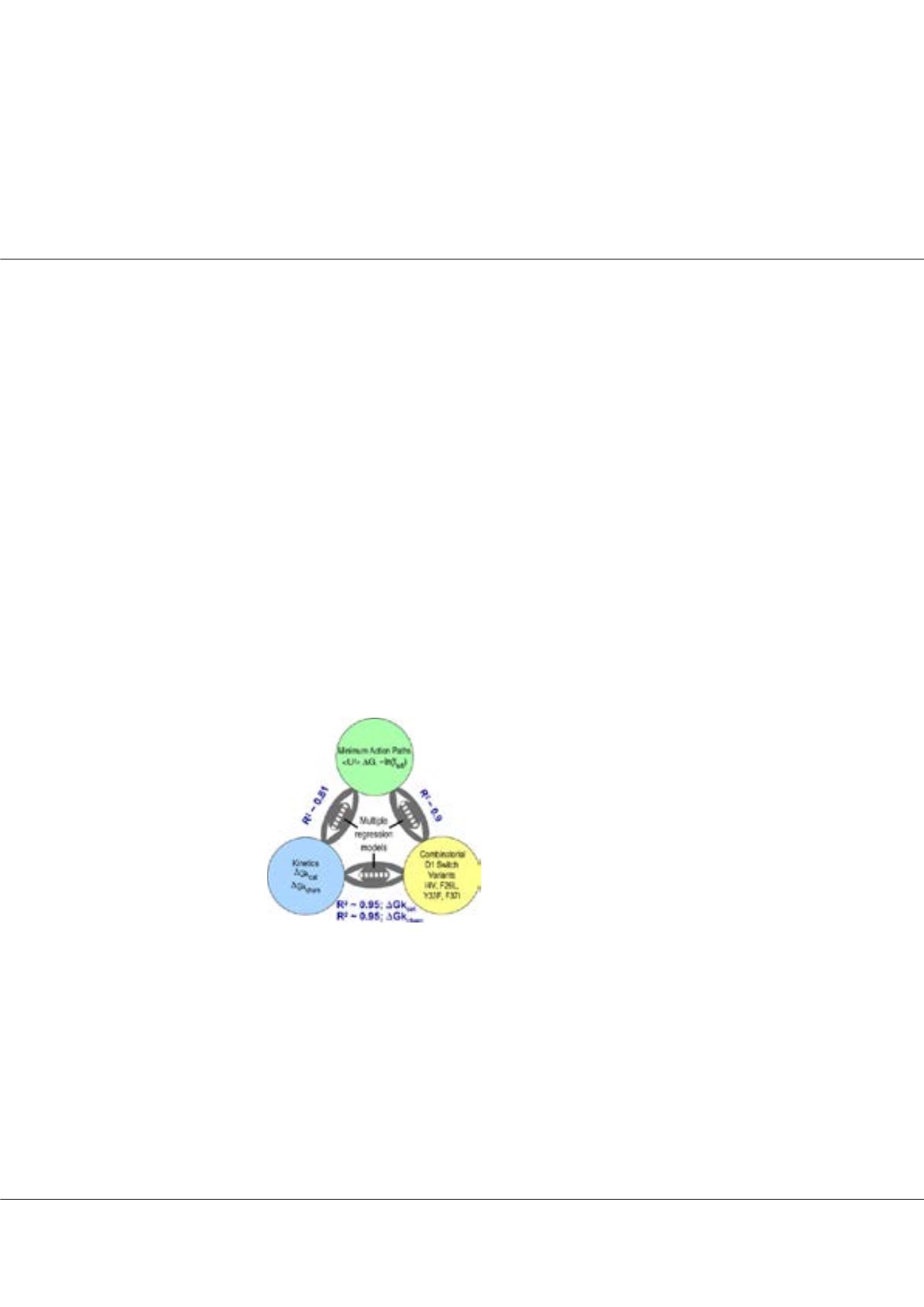
Figure1:
High Correlations between structures (yellow), steady-
state kinetics (blue) and computed trajectories (green) for WT
and 15 combinatorial variants of typtophanyl-tRNA synthetase.


















