
Page 140
Notes:
conferenceseries
.com
Volume 10, Issue 8 (Suppl)
J Proteomics Bioinform, an open access journal
ISSN: 0974-276X
Structural Biology 2017
September 18-20, 2017
9
th
International Conference on
Structural Biology
September 18-20, 2017 Zurich, Switzerland
Maharani Pertiwi Koentjoro et al., J Proteomics Bioinform 2017, 10:8(Suppl)
DOI: 10.4172/0974-276X-C1-0100
Crystal structure of DNA-binding domain-CbnR with its promoter reveals the basis of the LysR-type
transcriptional regulator recognition
Maharani Pertiwi Koentjoro
1
, Naruhiko Adachi
2
, Miki Senda
2
, Toshiya Senda
2
and
Naoto Ogawa
3
1
Gifu University, Japan
2
Institute of Materials Structure Science, Japan
3
Shizuoka University, Japan
C
upriavidus
necator
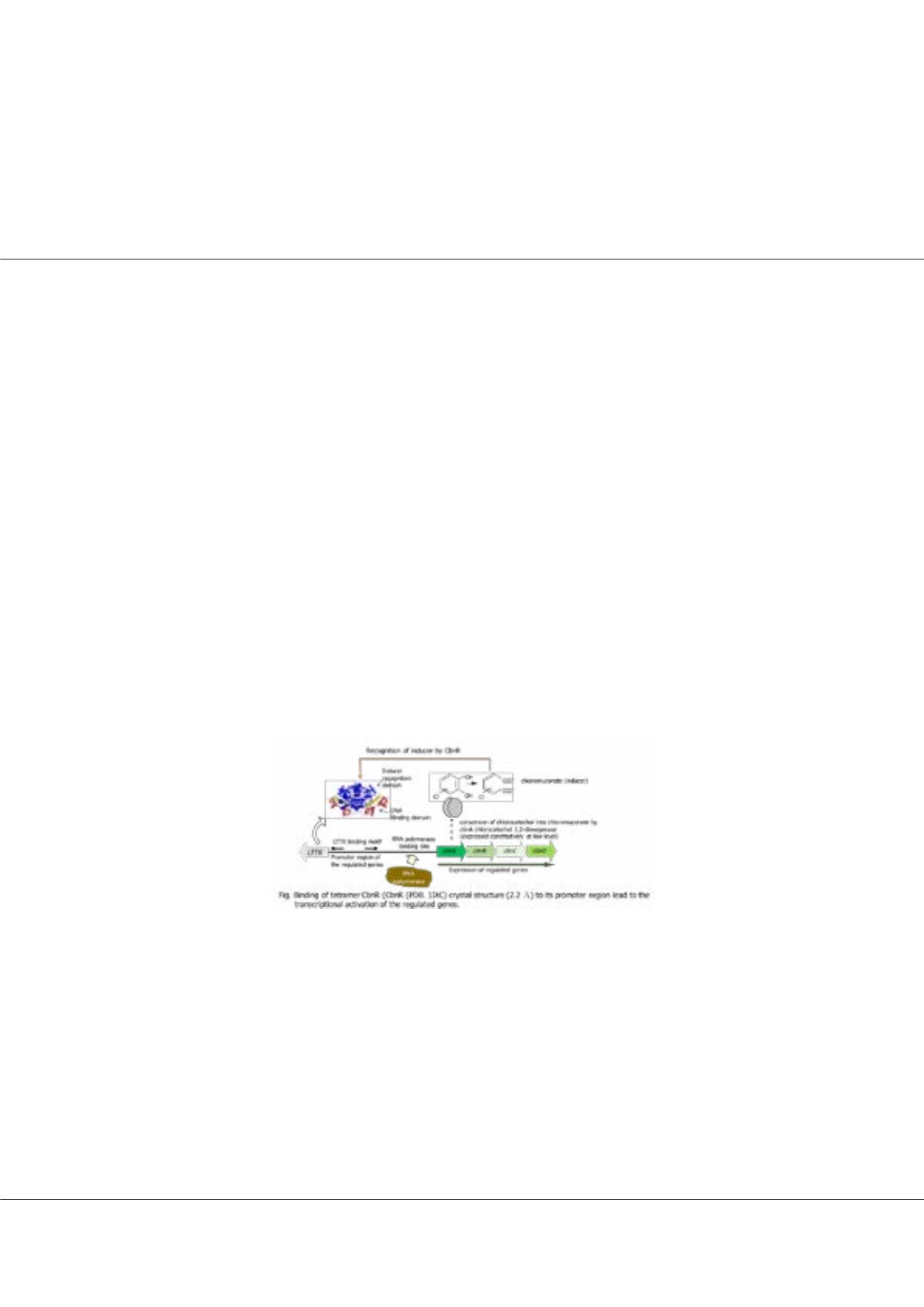
NH9, which can utilize chlorocatechol as a sole carbon and energy source, degrades chlorocatechol
with enzymes of the ortho-cleavage pathway. These enzymes are coded in the cbnABCD operon, of which expression is
specifically regulated by a LysR-type transcriptional regulator CbnR. CbnR forms a tetramer and can be regarded as a dimer
of dimers. The tetrameric CbnR has four DNA- binding domains and these DNA-binding domains recognize approximately
60 bp DNA sequence. The binding sequence is composed of two binding sites, recognition binding site and activation binding
site. Each binding site seems to be recognized by two DNA-binding domains in the tetramer. While the crystal structure of the
tetrameric CbnR has already been determined, the molecular mechanism of DNA recognition by CbnR remains elusive. We
therefore initiated the crystal structure analysis of DNA-binding domain of CbnR in complex with RBS. The crystal structure
would give an insight into the molecular mechanism of the CbnR-DNA interaction, which is the first step to understand the
gene activation mechanism by LTTR. Here we report the crystal structure of CbnR(DBD) (residues 1 - 87) in complex with
RBS, a 25-bp DNA fragment. The crystal structure was determined by the MR-native SAD method at 2.55 Å resolution with
Rwork/Rfree of 0.221/0.264.The crystal structure shows that dimeric CbnR(DBD) interacts with RBS.The dimeric CbnR(DBD)
adopts essentially the same conformation as that in the tetramic CbnR with the root mean squares deviation of 1.1 Å (174 Cα
atoms). The 3
α
helix and the winged region of the winged-helix turn helix motif in CbnR(DBD) directly interact with the
major and minor grooves of promoter sequence, respectively, and the interactions seem to bend DNA by approximately 30°.
To further analyse the molecular mechanism of their interaction, biochemical analysis is in progress.
Biography
Maharani Pertiwi Koentjoro is a 3
rd
year PhD student and the Monbukagakusho fellow in the United Graduate School of Agricultural Science, Gifu University, Japan.
She has completed her BA from Sepuluh Nopember Institute of Technology, Indonesia, and a Master in Gadjah Mada University, Indonesia. Her research interests
include molecular biological and biochemical investigation on bacteria. Currently she is working on structural studies of complex molecular machines that initiates
LysR-Type Transcription Regulator in bacteria.
maharanipertiwikoentj@gmail.com

















