Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
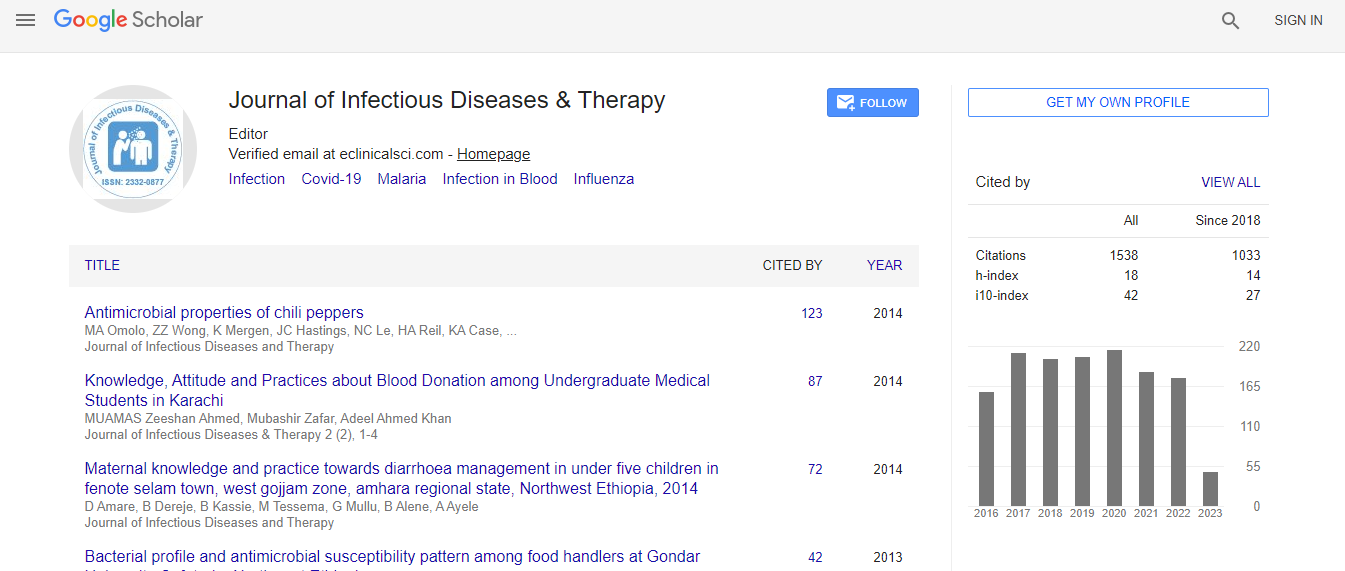
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Whole genome sequencing analysis from bacterial DNA: An attempt to Mycobacterium tuberculosis complete genome sequencing
3rd Annual Congress on Infectious Diseases
Alvarez-Maya Ikuri, Padilla-Martinez Felipe, Gonzalez-Barrios Juan Antonio, Barbadilla-Prados Antonio, Egea Raquel IBB and Islas-Rodriguez Alfonso
CIATEJ, Mexico Lara-Lozano Manuel Regional Hospital, Mexico Universitat Autonoma de Barcelona, Spain University of Guadalajara, Mexico
ScientificTracks Abstracts: J Infect Dis Ther
Abstract
Statement of the Problem: Tuberculosis is a bacterial disease caused by Mycobacterium tuberculosis. This bacterium is known for a high rate of drug resistance, and then tuberculosis is considered a worldwide public disease with high health and economic impact. Statistics in Mexico show that the incidence increases 15% every year, being a major problem due to the persistence. We aim to sequence the complete genome of Mycobacterium tuberculosis and subsequently perform bioinformatics analysis to determine possible molecular changes. Methodology & Theoretical Orientation: The complete genome of a Laboratory Mycobacterium tuberculosis strain H37Rv was sequenced using Next-Generation Sequencing (NGS) on the Illumina MiSeq platform. Genome DNA (gDNA) library was constructed using Nextera XT (Illumina) protocol. DNA was fragmented, tagged and selected by size, then sequenced by Illumina MiSeq-NGS platform. For bioinformatics, all sequences with adaptor contamination, duplicate reads or unknown nucleotides were removed by trimmomatic. Clean-filtered reads were mapped to the reference genome from GenBank (AL123456.3) by BWA software. Finally SAMTools software was used for SNP calling, since a resistance anti-tuberculous drug has been associated with SNPs in particular genes. Findings: Phred quality score in DNA sequencing was calculate (Q45) then this score was assigned to each nucleotide in the generated sequences. The P value was obtained (3.162e-005) and indicated that the genotype GC is very likely to be the true genotype in the sequenced sample. Preliminary results shown that there is a single nucleotide variant (SNV) from G to C at position 3982 in the strain of Mycobacterium tuberculosis. Conclusion & Significance: Mapping between Laboratory strain H37Rv and GeneBank H37Rv (ID 20829) shown at least one SNP in the position 3982. However, this result must to be confirmed using a higher depth reading and a further exhaustive analysis.Biography
Alvarez-Maya Ikuri is a Researcher at the Center for Research and Assistance in Technology and Design of the State of Jalisco. She holds a Post-doctoral degree in Neurobiology Department, NRC in University of Alabama at Birmingham UAB, Alabama, and USA; and in Department of Virology, Children's Hospital of Eastern Ontario CHEO, Ottawa, Canada. She has published in several indexed journals, attended more than 30 national and international congresses, and has contributed to the training of students in different levels of postgraduation. Her research interest is focused mainly on molecular diagnosis of infectious diseases.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi