Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
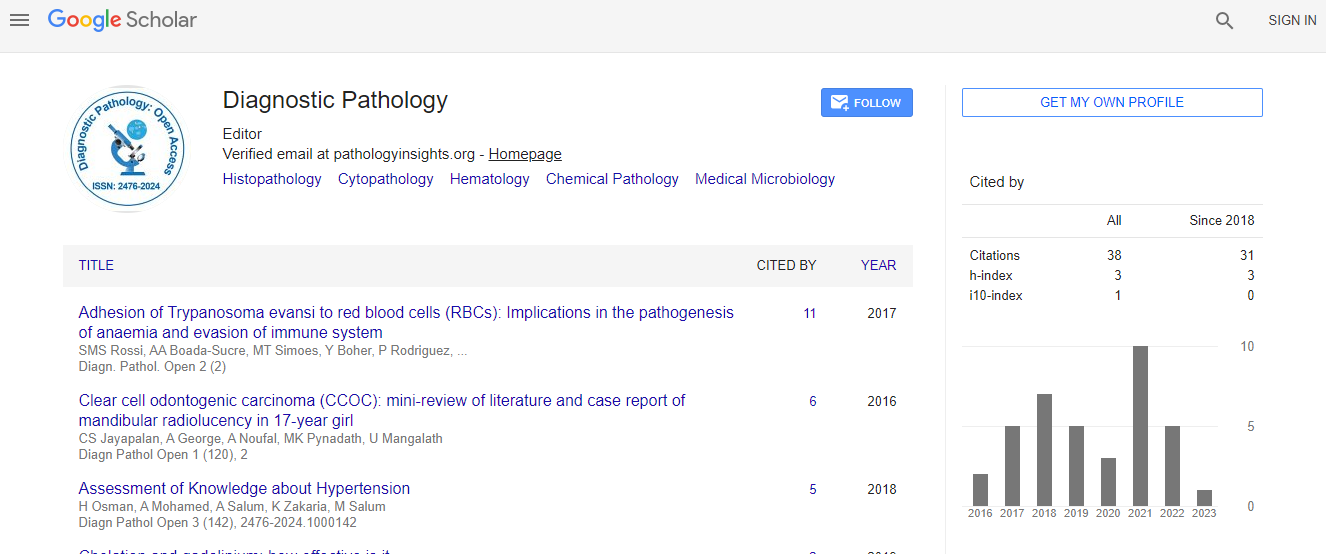
Google Scholar citation report
Citations : 110
Diagnostic Pathology: Open Access received 110 citations as per Google Scholar report
Diagnostic Pathology: Open Access peer review process verified at publons
Indexed In
- Google Scholar
- RefSeek
- Hamdard University
- EBSCO A-Z
- Publons
- Euro Pub
- ICMJE
Useful Links
Share This Page
Welcome to a labolution-The application of data mining and systems biology for personalized medicine
13th International Conference on Laboratory Medicine & Pathology
Christoph Hiemenz
PEPperPRINT GmbH, Germany.
ScientificTracks Abstracts: Diagn Pathol Open
Abstract
High-throughput technologies ranging from high-content microscopy screening to automated mass spectrometry pipelines and peptide microarray assays enable the generation of big medical data sets in a brief period of time with the requirement of very small analyte volumes. As a result, extremely large multiomics data sets can be generated which are ideally suited for statistical learning procedures using well established classifiers such as random forests or multivariate logistic regression models. To extract meaningful information from the data, a major challenge comprises the dimensionality reduction and unsupervised pattern recognition via machine learning algorithms including Fisher discriminant analysis, shrunken centroid analysis or clustering. Another problem to be tackled is represented by multivariate feature collinearities and ways have to be identified to remove this ambiguity. Yet, the reward for this mathematical rigor is the generation of reliable classifiers which can quickly stratify individual patients into disease/treatment subpopulations with acceptable sensitivity and specificity from multiomics data sets. Above all, technology suppliers such as opentrons and Oxford Nanopore Technologies promise the automated generation of multiomics data sets for a reasonable price.Biography
Christoph Hiemenz has completed his MSc from Ruprecht-Karls University Heidelberg. He work as a bioinformatician at PEPperPRINT GmbH and is the scientific instructor of the iGEM Team Heidelberg 2019. He has worked on diverse research projects: -DKFZ Heidelberg, Prof. Jörg Hoheisel- Kinase activity profiling with peptide arrays -BioQuant Heidelberg, Prof. Roland Eils/Prof. Barbara Di Ventura- Optogenetic nuclear protein shutteling and ODE modeling -KTH Stockholm, Prof. Stefan Ståhl – Surface functionalization of E.coli cells via sortases and inteins - EMBL Heidelberg, Dr. Carsten Schultz- Bioactivity of trifunctional Sphingolipids -IPMB Heidelberg, Prof. Andres Jäschke- Multivalent fluorescent turn-on probes for RNA imaging.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi