Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
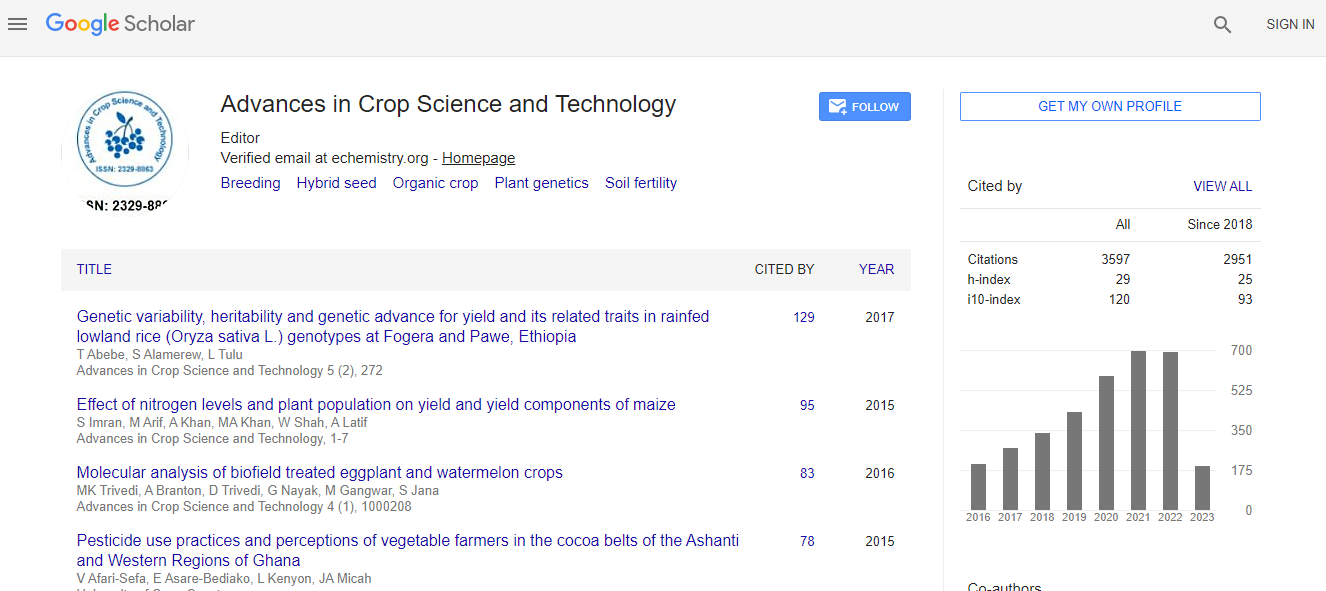
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
Transcriptome profiling for identifying genes to develop abiotic stress tolerant transgenic wheat
4th International Conference on Plant Genomics
Jasdeep Chatrath Padaria
National Research Centre on Plant Biotechnology, India
ScientificTracks Abstracts: Adv Crop Sci Tech
Abstract
Wheat (Triticum sp.) stands as the second largest staple crop of the world with 17% of the total cultivatable land under wheat production. Global annual wheat production needs to be increased at quantum leaps from the present production of more than 650 million metric tons so as to feed the ever burgeoning world population. Unfortunately, with the changing global climate, various abiotic stresses further hamper the wheat productivity. Development of abiotic stress tolerant cultivars is necessary to achieve the goal of enhanced wheat productivity. With the available gene pool within a species becoming limited, it becomes imperative that we search genes responsible for abiotic stress tolerance across the, genus, species and even kingdom and using rDNA technology develop transgenic wheat tolerant to abiotic stresses. The present study involved identification of abiotic stress responsive genes from tolerant plant systems as Penniseteum glaucum, Triticum aestivum, Ziziphus nummularia and Prosopis cineraria using Roche 454 and Illumina sequencing platforms. De novo assembly and transcriptome annotations were preformed to have insight about genes, gene family and transcriptional factors related to abiotic stress. Further analysis for change in expression level of known and unknown genes, SNP detection and SSR marker detection have been carried out to identify stress responsive genes and stress tolerance linked markers. A few stress responsive genes as NAC, P5CS, WRKY, HSP, MYB, ASR, DREB etc have been identified and characterized. These genes are further functionally validated and have been transformed in elite Indian bread wheat for development of transgenic wheat tolerant to abiotic stress.Biography
Jasdeep Chatrath Padaria has completed her PhD from Indian Agricultural Research Institute and Postdoctoral studies in the area of Gene expression profiling with respect to abiotic stress tolerance at Department of Horticulture and Landscape Architecture, Purdue University, USA. She is working as a Principal Scientist in the area of Biotechnology and Climate Change at National Research Centre on Plant Biotechnology, a premier institute in the area of plant molecular biology and biotechnology. She has published more than 25 papers in reputed journals and has guided 15 MSc and PhD students.
E-mail: jasdeep_kaur64@yahoo.co.in

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi