Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
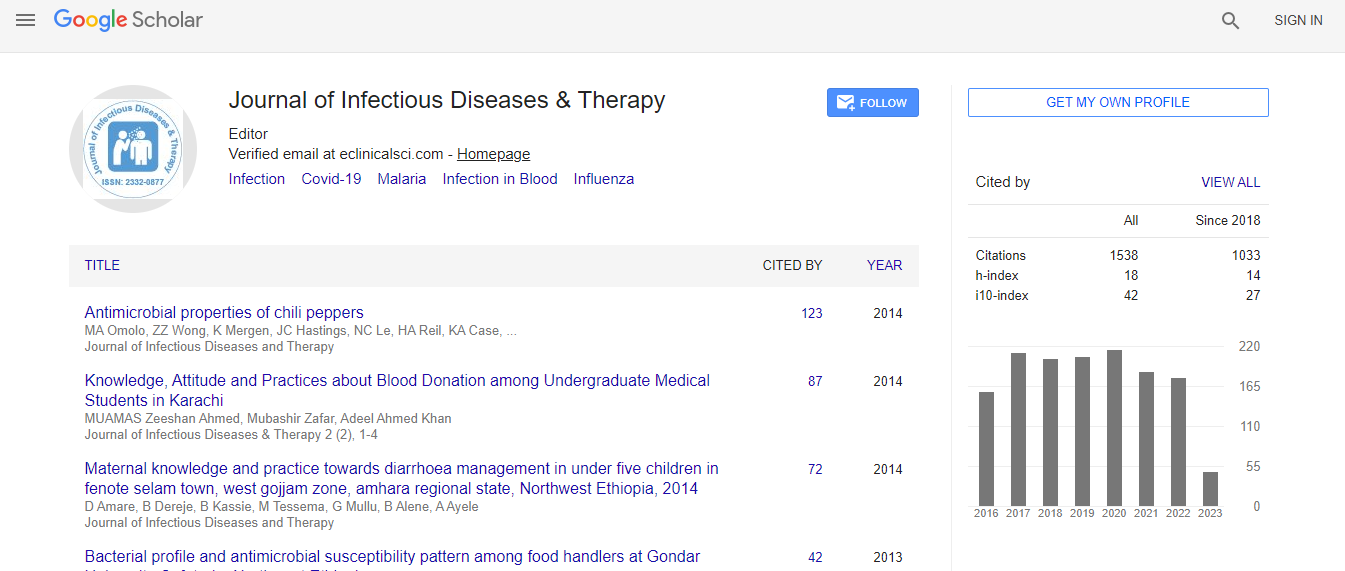
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Transcriptional signatures of Mycobacterium tuberculosis in mouse model of experimental intraocular tuberculosis
6th Euro-Global Conference on Infectious Diseases
Sudhanshu Abhishek, Michelle B Ryndak, Amod Gupta, Tulika Gupta, Shobha Sehgal, Suman Laal and Indu Verma
PGIMER, India NYC Medical Center, USA
ScientificTracks Abstracts: J Infect Dis Ther
Abstract
Statement of the Problem: Intraocular tuberculosis (IOTB), one of the extrapulmonary form of tuberculosis (TB), is a significant cause of inflammation and visual morbidity in TB endemic countries. Studies on IOTB are extremely challenging due to lack of appropriate human IOTB specimens, hence animal models of IOTB are required. Methodology & Theoretical Orientation: In the present study, a mouse model of IOTB was established by infecting the animals with Mycobacterium tuberculosis (M. tb; H37Rv) via intravenous (i.v.) route. Bacteriological evidence, histopathological changes and whole genome microarray study was done to identify the M. tb transcriptional signatures in mouse eye. Findings: At 45 days, post-infection (dpi), M. tb bacilli were observed in the eyes of 5 out of 12 (45%) M. tb challenged mice, whereas all the 12 animals showed positivity for M. tb RNA. Apart, histopathology of one CFU positive eye demonstrated intraretinal granuloma and moderate tissue damage in comparison to CFU negative eye that showed mild disease condition with no granuloma. Mycobacterium tuberculosis transcriptome analysis through microarray platform in the infected eyes, showed upregulation (ΓΆΒ?Β¥ 1.5- fold) of 12 M. tb genes, where top three upregulated transcripts included Rv0962c, Rv2612, and Rv0984. Real-time validation of these top three genes showed an average of 7.40, 4.13 and 3.47 Log2 fold upregulation (p<0.05), respectively. Conclusion & Significance: Although, ocular bacterial load was low, but detection of M. tb RNA with undetectable tubercle bacilli in the animals confirmed the paucibacillary nature of IOTB developed under experimental conditions, similar to that observed in human IOTB patients. Upregulation of mycobacterial genes, suggest that the adaptation of M. tb in ocular environment, an immuneprivileged site, may be associated with enhanced transcription of genes whose products are required for virulence and survival in intraocular environment. These genes/gene products could be important candidates for understanding the pathogenesis as well as development of new diagnostics/therapeutics for IOTB.Biography
Sudhanshu Abhishek has evolved from his Biotechnological skills and with Post-graduation in Human Genetics, to understand the infectious disease-like, Tuberculosis (TB). During his mid-tenure of PhD thesis, he was selected for NIH-FOGARTY Fellowships (USA), to be trained on Microarray Technology at NYU School Medicine, NY, USA. His keen evaluation and interest to understand the host-pathogen interaction has opened new avenues of research in intraocular TB through the models (animal and cell line), with a goal to understand the pathogenesis and early diagnosis of intraocular TB, which may lead to better therapeutics. He has grown well from his 6 years of Pre-doctoral training in this field through his continuous research, actively participating in teaching program of the department.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi