Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
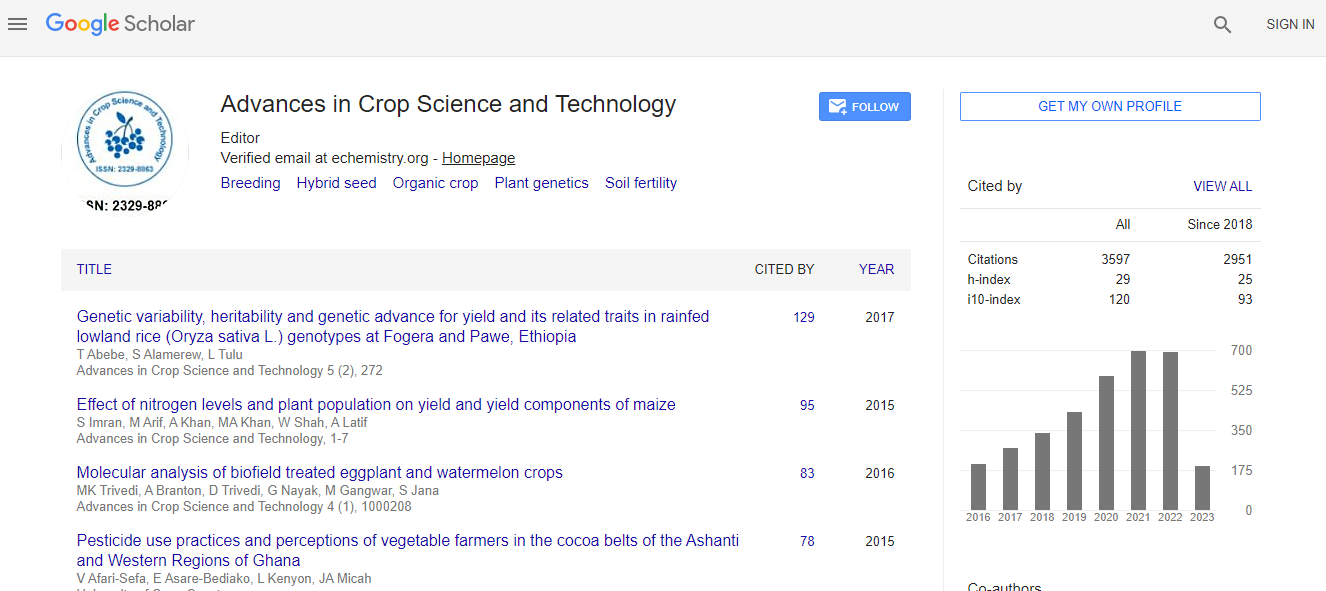
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
The use of DNA markers in plant breeding for disease resistance: From PCR-based approaches to allele specific markers
11th World Congress on Plant Biotechnology and Agriculture
Ahu Altinkut Uncuoglu, Yildiz Aydin and Ezgi Cabuk Sahin
Marmara University, Turkey
ScientificTracks Abstracts: Adv Crop Sci Tech
Abstract
Marker-assisted breeding is defined as the application of molecular markers in combination with linkage maps and genomics, to alter and improve plant traits on the basis of genotypic assays. Yellow rust, caused by Puccinia striiformis f.sp. tritici, is one of the major devastating factors worldwide in common wheat (Triticum aestivum L.). It is vital to identify associated DNA markers for Yr genes that can be used for marker assisted selection in wheat breeding programs to develop new cultivars with higher resistance. Here we report on the identification of six polymerase chain reaction (PCR)-based DNA markers (Xgwm382, Xgwm311, wmc658, PK54, BU099658, C6) linked with yellow rust resistance. Another issue requiring marker-assisted breeding are resistance for Plasmopara halstedii responsible for downy mildew disease and Orobanche cumana, holoparasitic plant called as sunflower broomrape, lead to loss of yield discount up to 100%. Single nucleotide polymorphism (SNP) markers linked with Pl8, Pl13 and Plarg resistance genes for downy mildew disease in combination with competitive allele specific PCR (KASP) assay which is a fluorescent tagged allele specific PCR method that is more efficient way to determine SNPs like insertions and deletions than other PCR techniques were identified. SNP markers (NSA2220 and NSA2251 for Pl8 gene, NSA0052 and NSA0354 for Pl13 gene, NSA2867 and NSA6138 for Plarg gene) were found discriminative among resistant and susceptible parents and their F2 populations. Also, evaluation of O. cumana races by KASP assay has been performed and SNP197 marker converted from the one SSR marker (Ocum-197), was found as a distinctive marker for O. cumana races. All these efforts mentioned above show the potential use and power of PCR-based and sequence-based DNA markers in plant breeding programs particularly for disease resistance in wheat and sunflower.Biography
Ahu Altinkut Uncuo├?┬?lu is currently working as Faculty of Engineering, Department of Bioengineering, Marmara University. She has completed her PhD at TUBITAK Marmara Research Center in 2001. Her research interest and specializations include biotic (plant diseases) and abiotic (drought and salt stresses) stress tolerance in crops at molecular level, molecular breeding, Marker Assisted Selection (MAS) studies in plant breeding, plant tissue culture and haploid plant production, association mapping and DNA barcoding studies in plants, understanding plant biodiversity at molecular level, technology transfer and university-industry relations in biotechnology area.
Email:ahu.uncuoglu@marmara.edu.tr

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi