Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
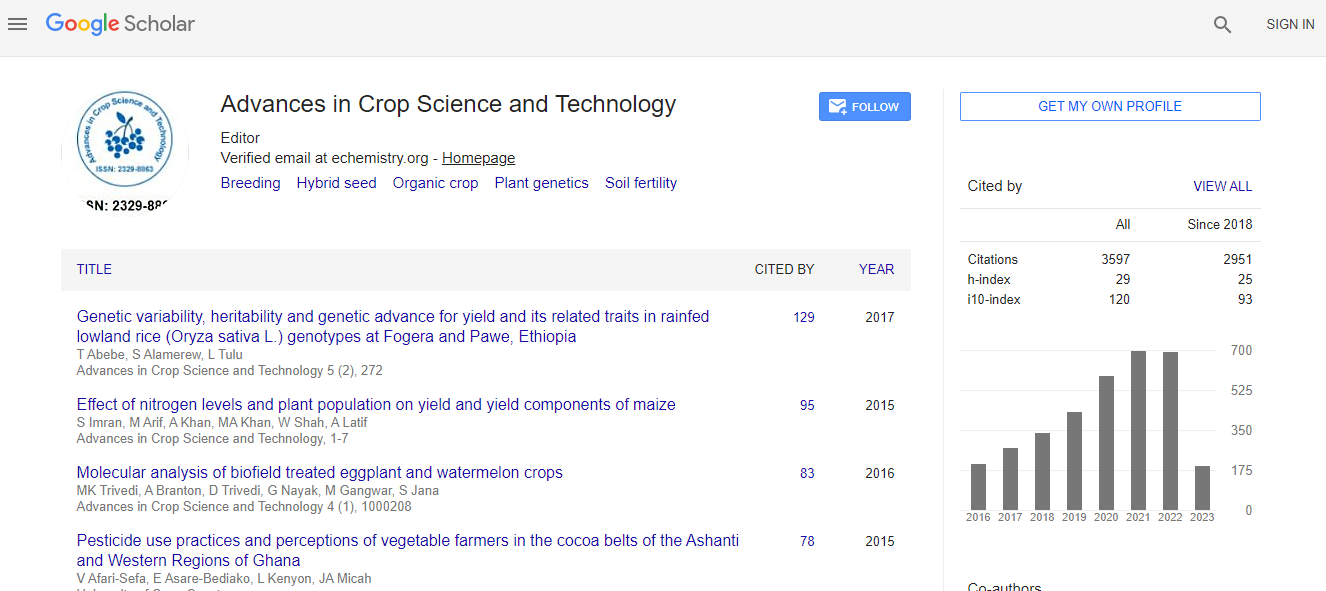
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
The rpg4/Rpg5 integrated decoy resistance to wheat stem rust race TTKSK in barley: Towards effector identification
4th International Conference on Plant Genomics
Robert Saxon Brueggeman, Roshan Sharma Poudel, Shyam Solanki, Shiaoman Chao and Jonathan Richards
North Dakota State University, USAUnited States Department of Agriculture-Agricultural Research Service, USA
ScientificTracks Abstracts: Adv Crop Sci Tech
Abstract
The barley rpg4/Rpg5 locus confers resistance against wheat stem rust caused by Puccinia graminis f. sp. tritici (Pgt) including race TTKSK (A.K.A. Ug99). The 70 kb region harbors two NLR R-genes, Rpg5 and HvRga1 that are required together for resistance. HvRga1 and Rpg5 contain typical NLR resistance-protein structure; however, Rpg5 has an additional C-terminal serine threonine protein kinase (STPK) domain. The transcription factor, HvVOZ1 was identified by yeast-two-hybrid of a library constructed from RNA of the rpg4/Rpg5+ line Q21861; 48 hours post inoculation, utilizing the Rpg5-STPK domain as bait. We hypothesize that the Rpg5-STPK acts as an integrated decoy that HvVOZ1 binds to negatively regulate defense activation or binds after activation as part of a signaling complex. The second NLR, HvRga1, may guard theHvVOZ1-Rpg5 interaction or surveil the Rpg5-STPK domain for Pgtrpg4/Rpg5-Avr (r45-Avr) effector manipulation. Thus, HvRga1 is possibly the guard that detects manipulation of the Rpg5 STPK or possibly HvVOZ1 by the r45-Avr effector eliciting a strong effector triggered immunity defense response. The r45-Avr needs to be identified to thoroughly investigate these mechanisms and test our hypothesis. To accomplish this a panel of 37 wheat stem rust isolates collected in North Dakota, many with differential race typing on the wheat differentials and differential reactions on rpg4/Rpg5 and Rpg1 in barley were genotyped using restriction site associated DNA-genotyping-by-sequencing (RAD-GBS). This RAD-GBS produced 4,919 informative SNPs and this initial genotyping was used to select 24 diverse isolates (16 avrRpg4/rpg5 and 8 Avrrpg4/Rpg5+) that were used to conduct in planta RNA-seq analysis during Pgt colonization 5 days post inoculation on the susceptible barley cultivar Harrington. The RNA-seq data was utilized to identify ~181,000 variant calls (SNPs and indels) within these Puccinia graminis transcriptomes during the infection process. The robust genotyping and phenotyping on these diverse differential isolates should allow us to identify candidate r45-Avr genes utilizing association mapping.Biography
Robert Saxon Brueggeman has completed his PhD in 2009 from Washington State University and Postdoctoral studies also from Washington State University Department of Crop and Soil Science. He is currently an Associate Professor at North Dakota State University as the Barley Pathologist/Molecular Geneticist. He has published more than 32 papers in reputed journals covering the topics of the cloning and characterization of barley disease resistance genes and fungal effectors.
E-mail: Robert.Brueggeman@ndsu.edu

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi