Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
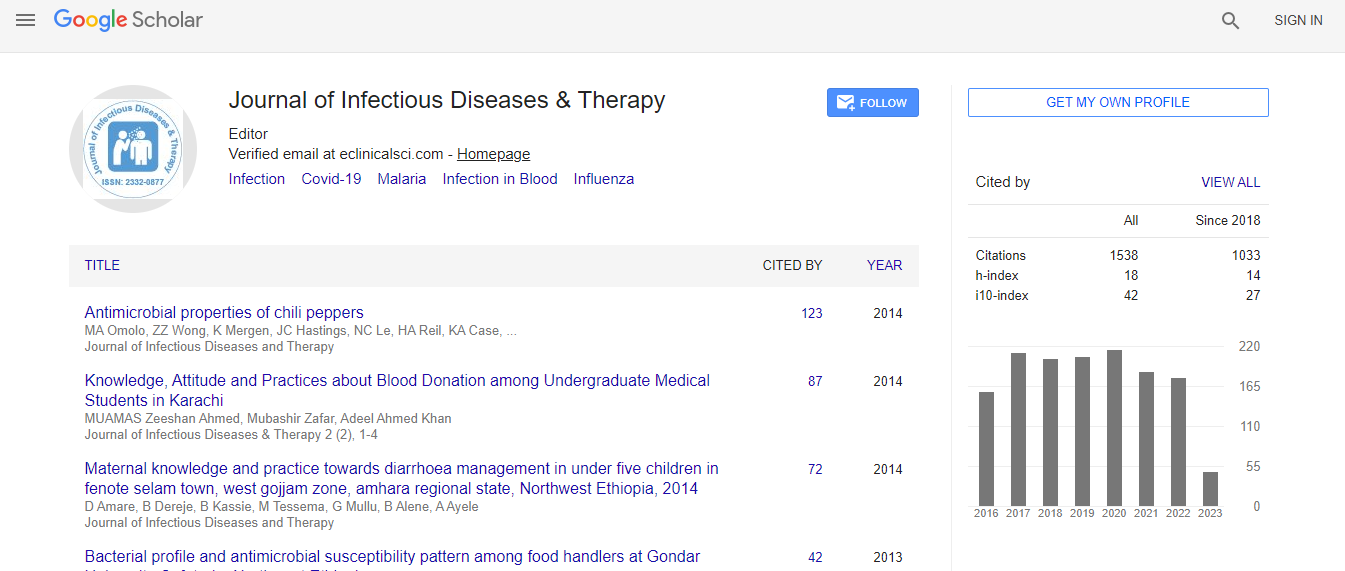
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
The neuraminidase universal epitope of influenza A virus induces a protective immune response in mice
2nd International Conference on Influenza
Zirak Javanmard Mahdieh, Taghizadeh Morteza, Moghaddam Pour Masoud and Arasteh Javad
Islamic Azad University of Tehran, Iran Razi Vaccine and Serum Research Institute, Iran
Posters & Accepted Abstracts: J Infect Dis Ther
Abstract
Human infection with influenza A virus is associated with a high mortality and morbidity and causes worldwide pandemics. There is essential to improve a universal vaccine against influenza pandemic. We identified a total of 12 conserved epitopes in viral neuraminidase proteins containing human T-cell epitopes for N1 & N2 subtypes. In this study, we use the epitope-based vaccine designed by immunoinformatics tools to predict the binding of B-cell and T-cell epitopes (class I and class II human leukocyte antigens [HLA]). BCPREDS was used to predict the B-cell epitopes. Propred, Propred I, netMHCpan and netMHCIIpan were used to predict the T-cell epitopes. The 3D molecular model was constructed by Swiss Model server and N-Glycosylation sites excluded from estimated regions. Important parameters like antigenicity and hydropathicity analyzed by Protean program. This sequence was cloned into the prokaryotic expression vector Pet-41b(+). BALB/c mice were immunized with different dosages of recombinant protein and the immune responses were determined in the form of protective response against influenza virus, antibodies titers, spleen cells lymphocyte proliferation and the levels of interferon-├?┬│ and interleukin-4 cytokines. We observed an increase in the number of influenza virus-specific IFN├?┬│-secreting splenocytes, composed of populations marked by CD4+ and CD8+ T cells producing IFN├?┬│ or TNF├?┬▒. Upon challenge with influenza virus, the vaccinated mice exhibited decreased viral load in the lungs and a delay in mortality. T-cells recognizing conserved epitopes were significant contributor to decreasing viral load and controlling disease severity during heterosubtypic infection in animal models.Biography
Email: mahdie.javanmard@yahoo.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi