Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
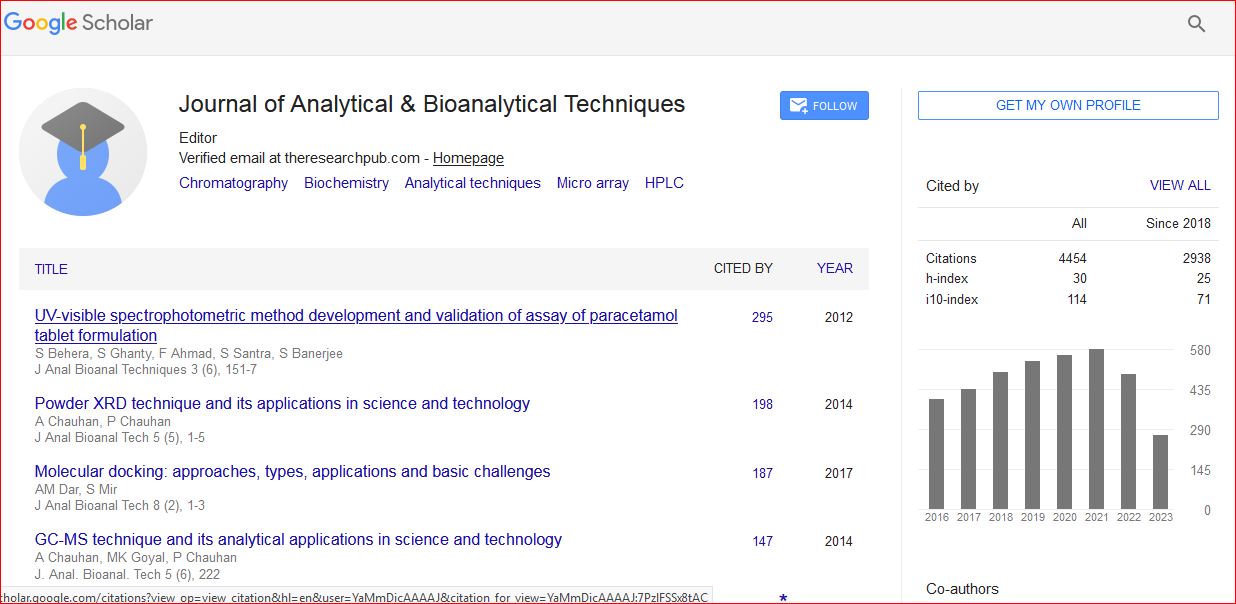
Google Scholar citation report
Citations : 6413
Journal of Analytical & Bioanalytical Techniques peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Euro Pub
- ICMJE
Useful Links
Related Subjects
Share This Page
Stuffer-free multiplex ligation-dependent probe amplification technology for identifying the copy number variation markers associated with the risk of SLE
6th International Conference and Exhibition on Analytical & Bioanalytical Techniques
Yeun-Jun Chung, Seon-Hee Yim and Seung-Hyun Jung
The Catholic University of Korea, Korea
Posters-Accepted Abstracts: J Anal Bioanal Tech
Abstract
Copy number variation (CNV) is one of the major components of human genetic variations and it is thought to contribute to inter-individual differences in diverse phenotypes. Several CNVs have been suggested to be associated with systemic lupus erythematosus (SLE) through the target gene approach; however, genome-wide feature of CNVs in the risk of SLE has not well studied. Recently, our group studied the genome-wide CNVs with 964 SLE patients and 711 normal control individuals by whole genome SNP array analysis, and identified three CNVs of RABGAP1L, 10q21.3, and C4 associated with the risk of SLE in Korean Women (Arthritis and Rheumatism 65:1055-63 2013). For the multiplex detection of the 3 CNVs, we adopted a stuffer-free multiplex ligation-dependent probe amplification based on conformation-sensitive capillary electrophoresis, which we developed previously (MLPA-CE-SSCP, Electrophoresis 33, 3052ΓΆΒ?Β?61 2012). For the MLPA-CE-SSCP based SLE CNV detection system, we also included 3 previously reported CNVs such as CCL3L1, FCGR3B, and EGR2. As a result, all the targets were well separated and the expected CNVs were consistently identified by the SLE MLPA-CE-SSCP system. After validation of the performance of SLE MLPA-CE-SSCP system, we applied the system for 113 SLEs and found that deletion of C4 was the most significantly associated with the risk of SLE. Our results suggest that the SLE MLPA-CE-SSCP system can be useful for predicting the risk of SLE. However, further larger scale study will be required.Biography
Yeun-Jun Chung is a Professor of Medical Microbiology and Head of Integrated Research Center for Genome Polymorphism (Catholic Medical College, Korea). He has studied genomics based personalized medicine for past twenty years. Since he founded IRCGP, he has studied Variomics and Oncogenomics using array-CGH, GWAS, NGS, MLPA-CE-SSCP and BI tools. He has published >80 research papers including Gastroenterology, Genome Res, Arthritis Rheum, Cancer Res, Bioinformatics, J Pathol etc. Currently he is an editor-in-chief of Genomics & Informatics (official journal of Korean Genome Organization) and an Editorial Board Member of Experimental & Molecular Medicine and World Journal of Gastroenterology.
Email: yejun@catholic.ac.kr

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi