Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
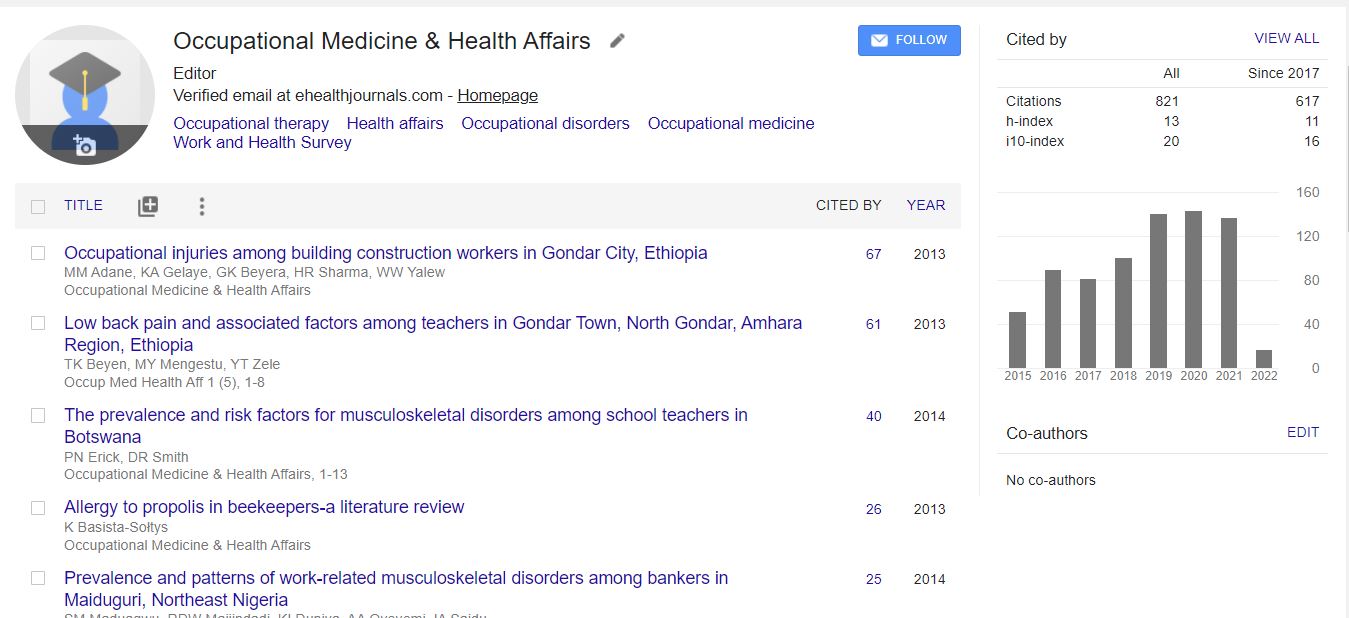
Google Scholar citation report
Citations :
Occupational Medicine & Health Affairs received citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- Academic Keys
- China National Knowledge Infrastructure (CNKI)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Geneva Foundation for Medical Education and Research
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
POLYCYCLIC AROMATIC HYDROCARBONS (PAHS): THE PECULIARITIES OF THEIR DEGRADATION BY LIGNINOLYTIC FUNGI
2nd International Conference on Environmental Health & Global Climate Change
Olga Turkovskaya, Natalia Pozdnyakova, Ekaterina Dubrovskaya, Sergei Golubev and Svetlana Balandina
Russian Academy of Sciences, Russia
Posters & Accepted Abstracts: Occup Med Health Aff
Abstract
Primary screening of 20 strains of ligninolytic fungi belonging to wood- and soil-inhabiting basidiomycetes and ascomycetes for degradative activity toward PAHs (phenanthrene, anthracene, and fluorene) showed that all the basidiomycetes examined were active in degrading the studied compounds. Of the three compounds tested, anthracene was the least available to all the fungi. The degradation of this compound varied from 16 to 91%. P. ostreatus MUT2977, Lenzites betulina, T. versicolor MUT3403, and T. maxima metabolized phenanthrene, anthracene, and fluorene more intensely (up to 90%). Phenanthrene and fluorene were degraded by basidio- and ascomycetes. Their decrease with P. ostreatus D1 was about 100%. One of the most active was the ascomycete C. herbarum, which degraded all the PAHs 100%. The ligninolytic enzyme system catalyzes the key stages of PAH degradation by fungi. We showed that all examined members of the genera Pleurotus and Trametes, as well as Len. betulina, St. murashkinskyi, and Sch. commune, produced laccase and Mn-peroxidase. Lignin peroxidase and Mn-peroxidase activities were detected in B. adusta only, whereas laccase activity was in Str. rugosoannulata only. These properties are typical of these fungi. We found for the first time that PAH degradation by the ascomycetes Lec. aphanocladii, F. oxysporum and C. herbarum was accompanied by the production of Mn-peroxidase only. In the ascomycete G. candidum, ligninolytic enzyme activity was not found. Despite some differences, the degradation of phenanthrene, anthracene, and fluorene followed the same scheme, forming quinone metabolites at the first stage: 9,10-anthraquinone in the case of anthracene, 9-fluorenone in the case of fluorene, and phenanthrene-9,10-quinone in the case of phenanthrene. Data were obtained which supported the hypothesis that the degree of PAH degradation may depend on the composition of the extracellular ligninolytic complex. The presence in the cultivation medium of only laccase (Str. rugosoannulata) resulted in accumulation of the corresponding quinones in the medium. Successive production of laccase and Mn-peroxidase (P. ostreatus D1) resulted in the formation and subsequent utilization of these metabolites. The simultaneous presence of two enzymes, the activity of one of which is low (St. murashkinskyi), resulted in slow degradation of these quinones. Finally, if both enzymes were highly active (T. hirsuta), the quinone metabolites formed were degraded quickly. The metabolites of �deep� degradation of the PAHs (2,2�-diphenic and phthalic acids), which are included in basal metabolism of fungi, were found. Therefore, the affiliation of the fungi with different eco-physiological groups and their cultivation conditions affect the composition and dynamics of production of the ligninolytic enzyme complex and, consequently, the completeness of PAH utilization. From the data obtained, we speculate that laccase can catalyze the initial attack on the PAH molecules to give quinones and that peroxidase catalyzes the following oxidation of these compounds, ultimately resulting in pollutant mineralization.Biography
Olga Turkovskaya is head of the Environmental Biotechnology Laboratory at the Institute of Biochemistry and Physiology of Plants and Microorganisms, Russian Academy of Sciences. Her Main research areas are degradation of xenobiotics by bacteria, plants, and fungi at natural sites and in wastewater, basic aspects of plant and microbial interactions with man-made environmental pollutants, influence of pollutants on natural ecosystems, selection and study of Plant-Growth-Promoting Rhizobacteria (PGPR) and development of biotechnologies for nature protection.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi