Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
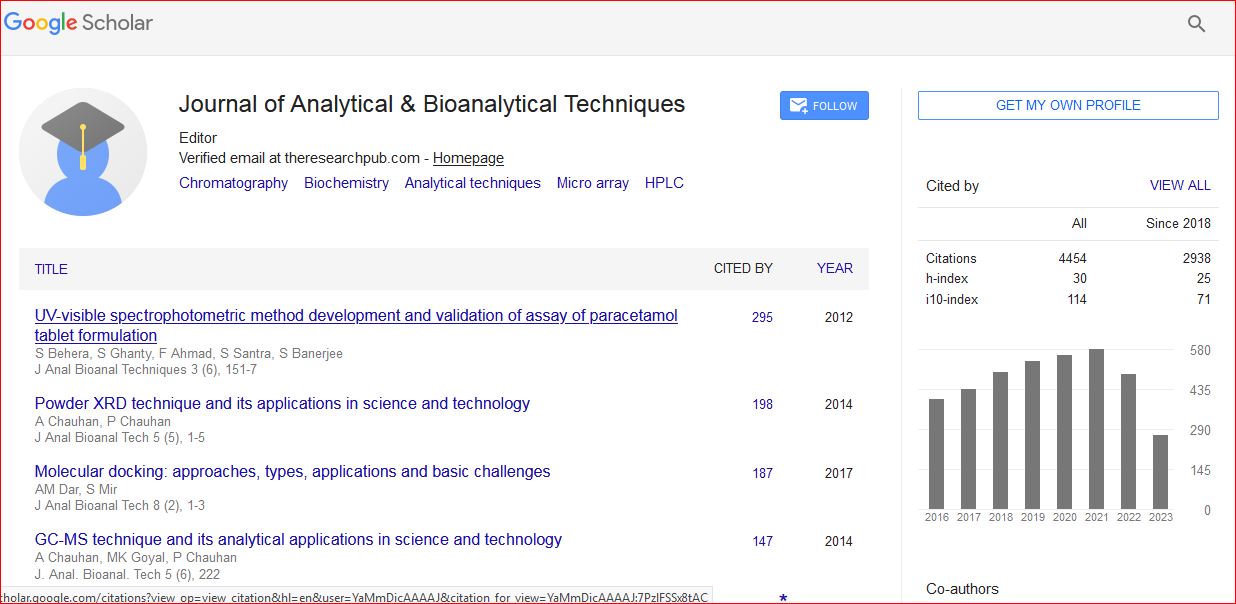
Google Scholar citation report
Citations : 6413
Journal of Analytical & Bioanalytical Techniques peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Euro Pub
- ICMJE
Useful Links
Related Subjects
Share This Page
PALMS: Parallel targeted/untargeted metabolome screening platform
7th International Conference and Exhibition on Analytical & Bioanalytical Techniques
Melnik A1, Da Silva R, Hyde E, Fernando Vergas, Amina Bouslimani, Ivan Protsyuk, Theodore Alexandrov and Pieter C Dorrestein
University of California, USA European Molecular Biology Laboratory, Germany Steinbeis Center SCiLS Research, Germany
Posters & Accepted Abstracts: J Anal Bioanal Tech
Abstract
The human body contains about 10 microbes per each human cell if you do not consider red blood cells which are lacking DNA. This means microbes significantly impact the chemistry in and on our body. Despite the role of microbial chemistry on human health, we know very little about its chemistry, let alone quantitatively. One of the analytical methods that are sensitive enough to measure microbial chemistry can be mass spectroscopy (MS). Here we introduce PALMS platform that is capable of analyzing full sample profile using high accuracy Orbitrap instrument and quantify set of targeted metabolites using triple quadrupole in a single LC run. Several hundred of fecal swabs samples from the American Gut project were tested and then people were swabbed at several hundred locations for the creation of 3D quantitative maps. The system setup comprises of triple quadrupole (qqq) and Orbitrap mass spectrometers connected in parallel to a single UHPLC system. Liquid connection was split into equal ratio and directed into two MS which are equipped with identical HESI ion sources; tubing length maintained equal in order to avoid any retention time (RT) differences between two MS. The data acquisition was triggered by UHPLC system by mean of contact closure signal that is sent to both MS simultaneously. Data captured by Q Exactive recorder in untargeted profiling mode with high resolution while quantitative data on defined targets were acquired by TSQ. We first evaluated our system by screening of several hundreds of fecal swabs extracts received from American Gut project. For targeted quantification we used mix of 200 standards containing microbial primary and secondary metabolites. We evaluated a distribution of the standards in fecal samples by constructing a calibration curve with triple quadrupole data. Our results showed that even in such high background as fecal matter we can quantify metabolites with relatively low error rate. By feeding mass list to triple quadrupole we managed to achieve pictogram sensitivity at the same time acquiring a full molecular profile using Orbitrap. By analyzing untargeted data we were able to putatively identify thousands of molecules using molecular networking while quantification was performed for selected microbial metabolites. Because in molecular networking structurally similar molecules clustered together we were able to relatively quantify analogs of the standards by extrapolating their concentrations. We then used the platform for screening the microbial metabolites reside on human body. We constructed 3D chemical composition maps of human skin surface. Highly accurate, untargeted data allowed for identification of some key microbial metabolites that are distributed on the skin in the specific locations such as armpit and groin area. We then relatively quantified the metabolites as some of our standards clustered with molecules of interest, which concentration was accurately measured by triple quadrupole. Determining the quantitative information for key microbial metabolites grant us the possibility to calculate the amount of the microbial cells that are primary producers and potentially assess the impact of them on our health.Biography
Email: alexvmelnik@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi