Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
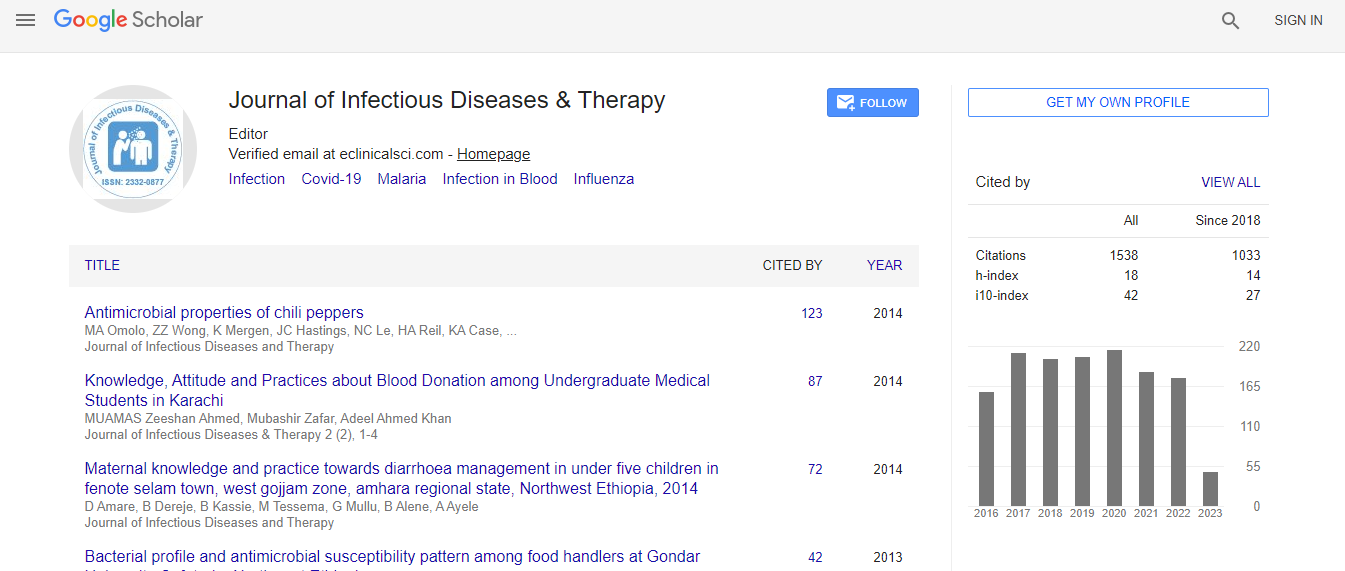
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Monitoring influenza viruses√ʬ?¬? genetic data: A touchstone in understanding the virus-host interaction and in selection of seasonal influenza vaccine√ʬ?¬?s strains
International Conference on Influenza
Mihaela Lazar1, Alina Elena Ivanciuc1, Carmen Maria Cherciu1, Gheorghe Necula1, 4, Daniela Pitigoi1,2, Odette Popovici3, Rodica Popescu3, Maria Elena Mihai1, Cristina Tecu1 and Emilia Lupulescu1
1National Institute for Research √ʬ?¬?Cantacuzino√ʬ?¬Ě, Romania 2University of Medicine and Pharmacy Carol Davila, Romania 3National Institute of Public Health, Romania 4HoriaHulubei√ʬ?¬Ě National Institute for R&D in Physics and Nuclear Engine
Posters-Accepted Abstracts: J Infect Dis Ther
Abstract
In Romania, the influenza activity in 2014-2015 was characterized by co-circulation of influenza A(H3N2), A(H1N1) pdm09 and B viruses. Primary objective of our study was to monitor the emergence of HA variants of influenza virus in patients with influenza-like illness (ILI) and severe acute respiratory infections (SARI) selected from sentinel and non-sentinel systems by antigenic and genetic characterization. One hundred eleven strains were isolated on cell cultures. All A(H1N1) pdm09 viruses were antigenically characterized as A/California/7/2009 but a higher heterogeneity was observed for subtype H3N2 and type B indicating a mismatch between circulating and vaccine strains. Full hemagglutinin gene was sequenced from 22 original specimens (including an influenza B and H1N1 co-infection), from which 14 collected from patients hospitalized with SARI. Four of them had fatal evolution, although one had been vaccinated. All characterized viruses fell into clade A/California/7/2009, subgroup 6B, A/South Africa/3626/2013. Seventeen viruses type B fell into clade 3 (B/Phuket/3073/2013), four from vaccinated patients and three deaths. For A(H3N2) viruses, the complexity of genetic data interpretation must take into account the nature and location of the main mutations. The 22 sequenced strains H3N2 clustered into three different clades, 3C.2a and 3C.3a and 3C.3b. However, among deaths with laboratory-confirmed influenza, only 15% were caused by the H3N2 subtype versus 62.5% subtype H1N1 and 22.5% type B respectively. Our study emphasize the importance of monitoring viral evolution and the role of genetic data on understanding the virus-host interaction and disease severity as well as in selecting seasonal influenza vaccine√ʬ?¬?s strains.Biography
Mihaela Lazar is a Researcher and main player in the National Surveillance Program of ILI (Influenza-like Illness) and SARI (Severe Acute Respiratory Infections) network in Romania.
Email: mlazar@cantacuzino.ro

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi