Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
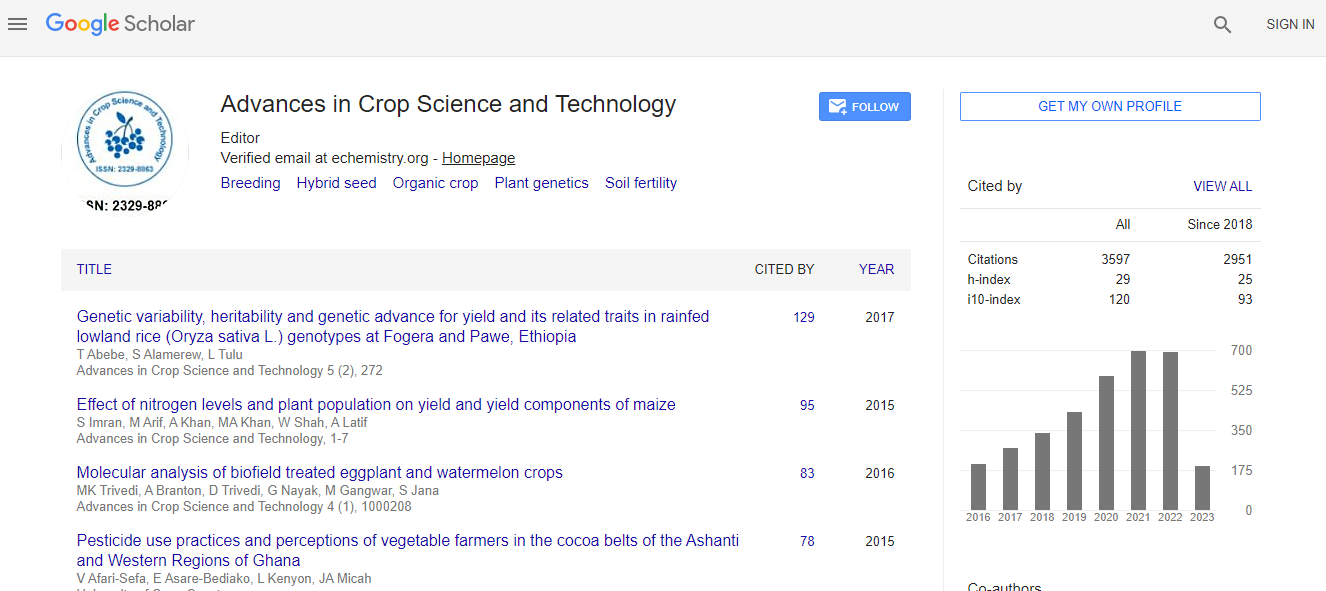
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
Molecular characterization of ten Heliconia (Heliconiaceae) genotypes by means of RAPD markers
11th World Congress on Plant Biotechnology and Agriculture
Moumita Malakar, Pinaki Acharyya and Sukanta Biswas
University of Calcutta, India
ScientificTracks Abstracts: Adv Crop Sci Tech
Abstract
Heliconia a trendiest specialty cut flower of tropics is securing popularity for its inflorescence��?s charismatic form and alluringly blended hues in the world and in Indian market too. It is a monophyletic genus with roughly 200 to 250 species of neotropical origin. It resides alone in Heliconiaceae family of Zingiberales order former incorporated in Musaceae for fusion of inverted flowers, single staminode and drupe type fruits elite for it. Exploitation of molecular markers may aid in perceptive recognition of several species and varieties of them since taxonomic confusions and uncertainties subsist amid them. In this context, ten species and varieties were analyzed using RAPD markers. Chosen thirty primers among seventy, amplified 1281 polymorphic DNA fragments with each primer giving a mean of 42.7 polymorphic bands. The genetic similarity matrix constructed with Jaccard��?s coefficient using RAPD marker scores showed that the highest value was between H. psittacorum var. Choconiana and H. psittacorum var. Lady Di (0.384), while the lowest was between H. psittacorum var. Choconiana and H. stricta var. Dwarf Jamaican Red (0.244). Ten species and varieties of Heliconia formed three distinct clusters at similarity coefficient value of 0.33, implying a parallelism between genetic and morphologic or taxonomic variability of Heliconia genotypes. Golden Torch, Choconiana, Lady Di and H. wagneriana included in cluster I while Choconiana, Lady Di formed a more cohesive entity. On the contrary, H. stricta var. Dwarf Jamaican Red produced a separate cluster validating that taxonomically related entries clustered together and distant ones segregated. So, the current sequels demonstrate that RAPD is a rapid, relatively economical and functional for the characterization of genetic divergence between different Heliconia genotypes.Biography
Moumita Malakar is a Senior Research Fellow (INSPIRE Fellow) at University of Calcutta, India, pursuing Doctoral program on Horticulture, specializing in Floriculture & Landscaping. She studied longevity of few ornamental foliages and flowers.
Email:moumitamalakar7@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi