Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
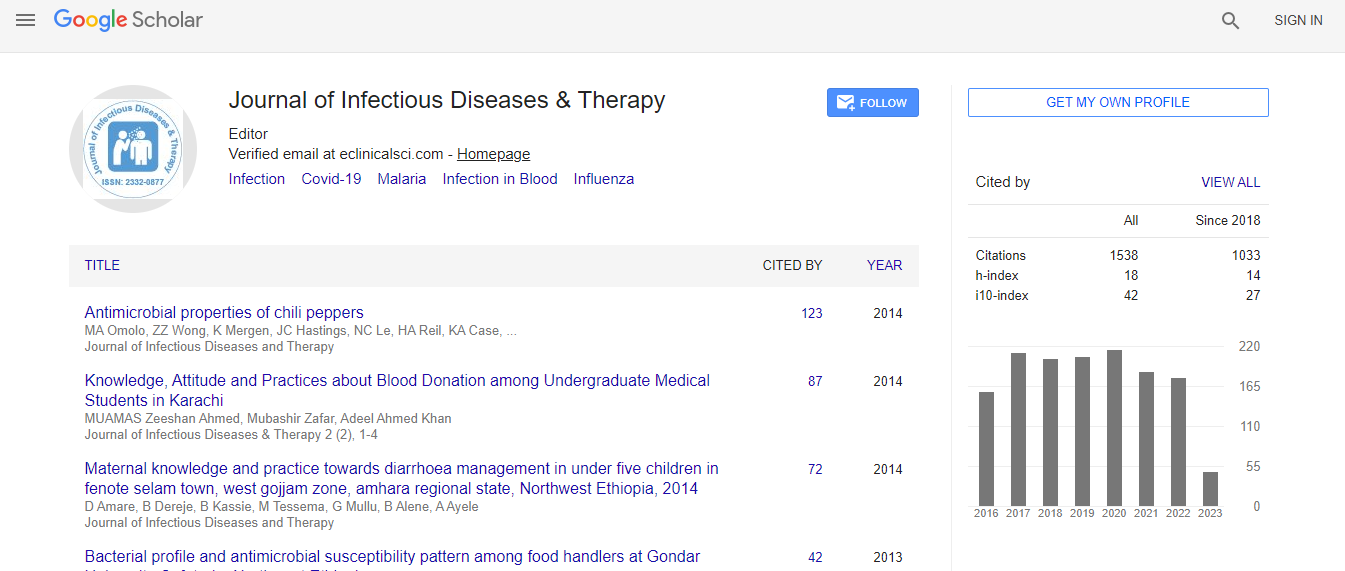
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Molecular characterization of influenza viruses among infants and children in Casablanca - Morocco during four post-pandemic seasons: 2010-2014
International Conference on Influenza
L Anga1, A Faouzi1, L Benabbes1, A Idrissi2 and J Nourlil1
1Pasteur Institute, Morocco 2Ministry of Health, Morocco
Posters-Accepted Abstracts: J Infect Dis Ther
Abstract
Background: Influenza remains a major health problem due to the annual seasonal epidemics caused by the emergence of new strains. Hemagglutinin (HA) and neuraminidase (NA) glycoproteins are submitted to frequent changes by antigenic drift. Children are more likely to get the flu or have flu-related complications mostly with chronic health conditions such as asthma. Our main objective was to identify the influenza viruses among children during four post-pandemic seasons 2010-2014. Other respiratory viruses such as respiratory syncytial virus, rhinovirus and adenovirus were detected. Methods: Nasopharyngeal samples were collected from infants and children and analyzed in our laboratory. The isolation of virus per inoculation was done on MDCK cells and the viral identification was performed by Real time PCR (qPCR) and nucleotide sequencing targeting the matrix, HA and NA genes, according to protocols recommended by CDC Atlanta and NRC for Influenza (IP Paris).The nucleotide sequences were analyzed and compared with sequences of the vaccine virus strains available in the same season of sample collection. Results: A total of 552 samples from infants and children aged between 11 days and 15 years with respiratory illness were collected between week 45/2010 to week 10/2014. The following viruses were detected: 58 (10.5%) influenza A/H1N1pdm09, 77 (14%) A/H3N2, 49 (8.87%) influenza B, 68 (12.31%) RSV-A, 5 (1%) RSV-B, 24 (4.43%) rhinovirus and 7 (1.26%) adenovirus. Co-infections were also identified: A/H1N1pdm09 and B (2 cases), rhinovirus and RSV (9 cases), adenovirus and influenza B (1 case), A/H1N12009 and adenovirus (1 case). Based on the phylogenetic analysis, the HA1 region gene of both H1N1pdm09, H3N2 isolated subtypes and HA1 region gene of influenza B clustered with the current vaccine strains: A/California/7/2009 (H1N1)-like (2010-2014), A/Perth/16/2009 (H3N2)-like for (2010-2012); A/Victoria / 361/2011 (H3N2)-like (2013-2014); B/Brisbane / 60/2008-like (2010-2011); B/Wisconsin/1/2010 (2012-2013). No known mutation conferring resistance to NA inhibitors such as H275Y was observed in any of the sequenced H1N1pdm09 isolates. Conclusion: During the study period, Moroccan influenza viruses were well matched with the Northern Hemisphere vaccine formulation. Surveillance of influenza viruses is important not only for development of national prevention and control strategies but also for earlier identification of the newly emerged strains. Further studies will be needed to investigate other respiratory viruses.Biography
Email: angalatifa@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi