Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
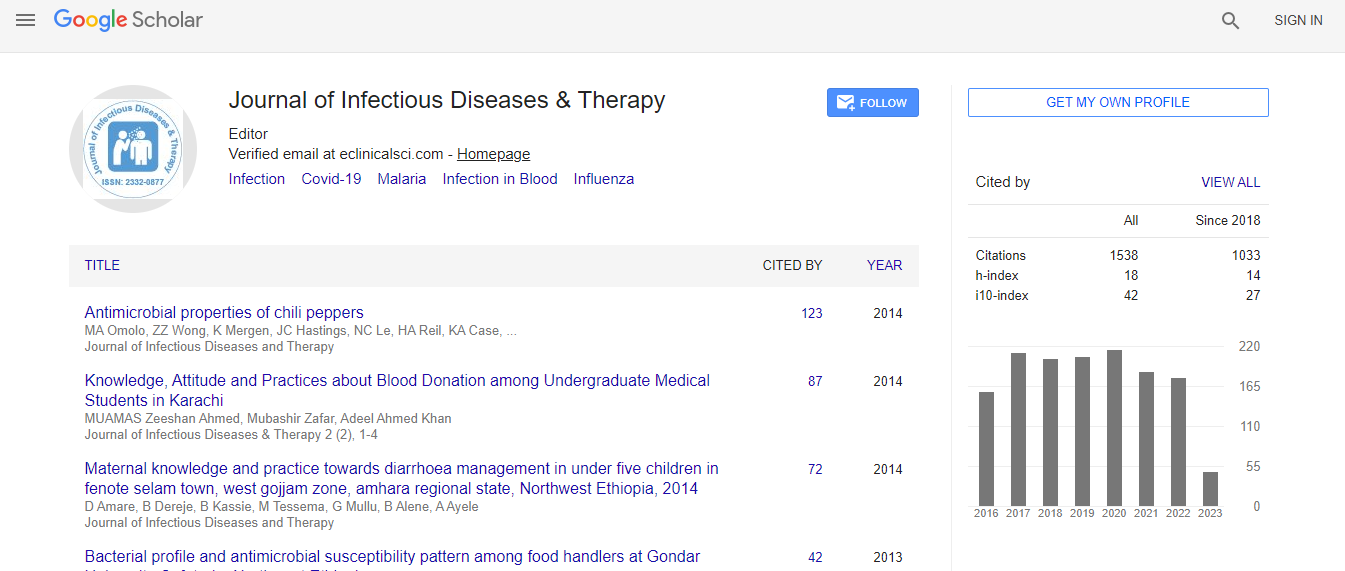
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Genotype shift of dengue-2 virus isolated in the Philippines revealed by whole genome analysis
Joint Event on 2nd World Congress on Infectious Diseases & International Conference on Pediatric Care & Pediatric Infectious Diseases
Maria Luisa G Daroy
St. Luke├ó┬?┬?s Medical Center, Philippines
ScientificTracks Abstracts: J Infect Dis Ther
Abstract
Whole genome analysis of dengue virus strains isolated in the Philippines was performed using an Illumina MiSeq next generation sequencer and bioinformatics. Complete genome sequences of 24 DENV-2 isolated from 1995 to 2008 was mapped against a DENV-2 reference sequence (NC_001474) from Genbank. The Philippine DENV-2 isolates were highly similar to strains from Taiwan (AJ968413.1) and China (EF051521.1). The Chinese strain (EF051521.1) was isolated in 2001 yet similar strains could have been circulating in the Philippines as early as 1998. The isolates highly similar to the Taiwanese strain were of the Asian II genotype and the isolates highly similar to the Chinese strain were of the Cosmopolitan genotype. In total, 922 synonymous and non-synonymous substitutions were observed, of which 232 (25.2%) were in the NS5 gene, which had the most substitutions. This was followed by NS3 with 181 (19.6%) and the E gene with 144 (15.6%) and the 2K gene with only 6 (0.7%). Putative amino acid sequences from the coding regions revealed that majority (817, 88.6%) of the substitutions were synonymous or silent. Of the remaining 105 non-synonymous mutations, 22 (20.9%) were in the NS5 gene followed by the NS1 and E genes with 20 (19.0%) and 16 (15.2), respectively. When the clustered genetic variation profiles were identified based on the specific clinical diagnosis, all the DHF III cases belonged to a single cluster together with some DF cases and one SVI case. In contrast, all DHF I-II and most of the DF cases grouped together in another cluster. All DHF III cases were of the Cosmopolitan genotype. DENV circulating within the Philippines from 1995 to 1998 were of the Asian genotype, followed by a period from 1998 to 2001 in which both the Asian II and Cosmopolitan strains co-circulated and from 2000 to 2008 the circulating DENV was primarily cosmopolitan. This supports previous observations of a probable genotype shift in the Philippines. This is the first report of whole genome analyses of dengue virus isolates from the Philippines.Biography
Maria Luisa G Daroy is a Scientist at the Research and Biotechnology Division of St. Luke’s Medical Center and Assistant Professor in the MS Molecular Medicine Program of the St. Luke’s College of Medicine-WHQ Memorial. She has published more than 20 papers on dengue, Japanese encephalitis, chikungunya, eye infections, dementia, diabetes and coronary artery disease. She was the Chair of the Board of Examiners of the Philippine Academy for Microbiology from 2013-2015 and authored a book chapter on Philippine microbiology research. Researches include dengue, chikungunya, diarrhea, CNS infections, pathogen genomics, antimicrobial resistance, plant antivirals, molecular diagnostics and genetics of CVD, thyroid cancer and dementia.
Email: mlgdaroy@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi