Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
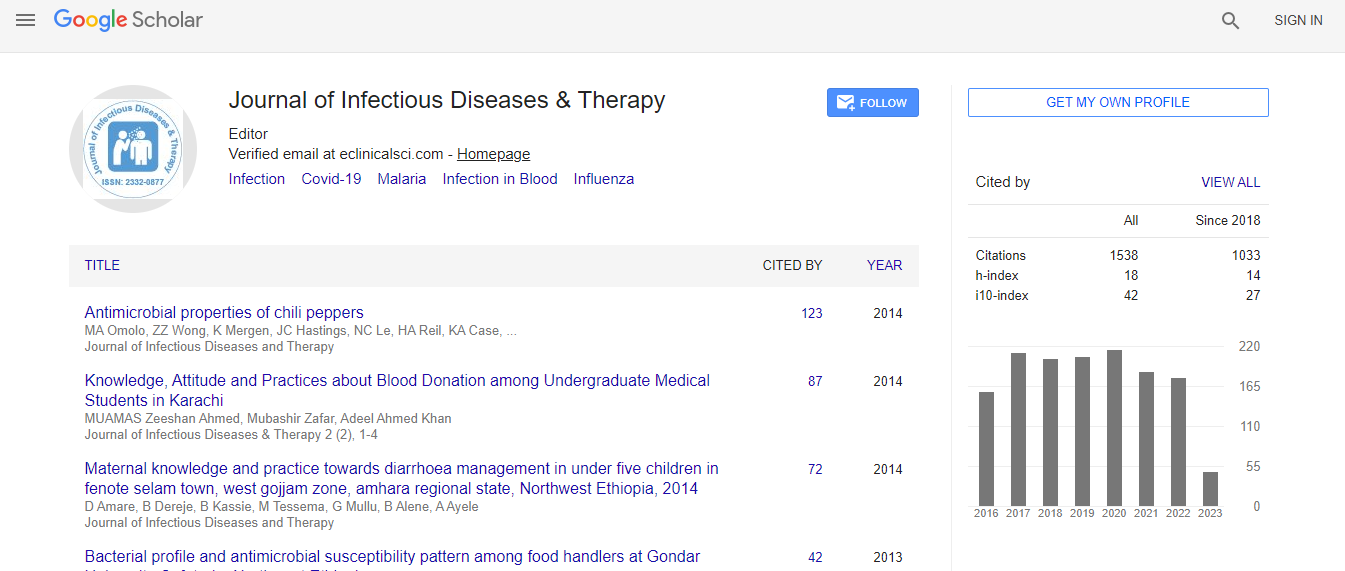
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Genetic diversity and amino acids variations at vaccine target sites in rabies viruses collected from different host species in Makueni and Siaya counties, Kenya
Joint Event : 10th International Congress on Infectious Diseases & 12th International Conference on Tropical Medicine and Infectious Diseases
Evalyne Wambugu*, Kimita Gathii, Sarah Kituyi, Michael Washington, Clement Masakhwe, Lucy Mutunga, Gurdeep Jaswant, SM Thumbi, Brian Schaefer and John Waitumbi
University of Embu, Kenya United States Army Medical Research Unit, Kenya Uniformed Services University, USA University of Nairobi, Kenya
Posters & Accepted Abstracts: J Infect Dis Ther

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi