Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
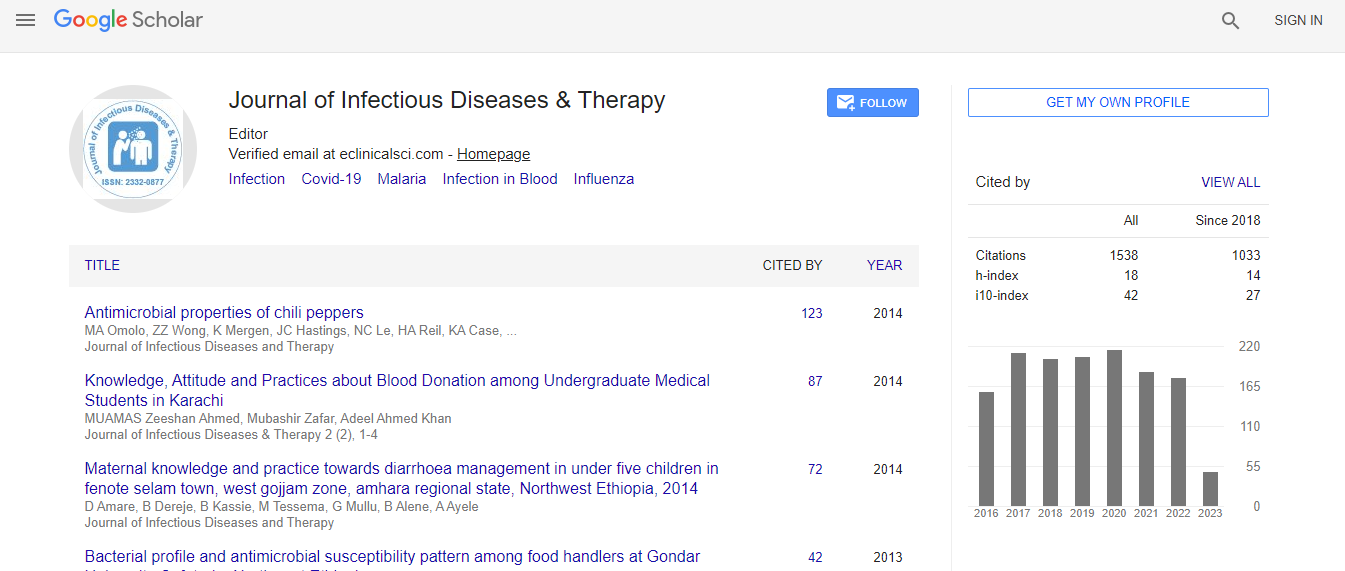
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Genetic characterization of co-circulating influenza A viruses in the pandemic (2009-2010) and post pandemic (2010-2011) periods in Mumbai
International Conference on Influenza
Devanshi Gohil, Sweta Kothari, Pramod Shinde, Rajas Warke, Abhay Chowdhary and Ranjana Deshmukh
Haffkine Institute for Training, Research and Testing, India
ScientificTracks Abstracts: J Infect Dis Ther
Abstract
Pandemic influenza A (H1N1) 2009 virus was first detected in May 2009 initiating the pandemic in India. Influenza A viruses has the ability to evade the immune response through the acquisition of genetic changes. Understanding the multiple lineages of influenza virus variants is necessary in lieu of co-circulation of influenza A (H1N1) pdm 09 and seasonal influenza A (H3N2) virus in Mumbai, a global transition hub. However, genetic information of circulating influenza strains in Mumbai is limited. In the present study, we performed molecular and evolutionary analysis of these co-circulating viruses isolated from patients during August 2009 ΓΆΒ?Β? March 2011. Positive samples were cultured and the viral genomes for HA and NA were analyzed for both seasonal and pandemic isolates. Molecular analysis revealed substantial sequence variations in both pandemic and seasonal influenza A viruses. HA1 sequences of pandemic isolates were conserved at the receptor-binding site, while variations in the amino acid substitutions at the antigenic sites resulted in the changes at the N-linked glycosylation sequon. In pandemic isolates, amino acid substitutions in the NA gene were dissociated with the catalytic or framework sites. Seasonal Influenza A virus exhibited antigenic drift. Phylogenetic analysis of the HA and NA genes of pandemic and seasonal isolates illustrated their evolutionary closeness or drift with their concomitant prototype vaccine strains respectively. Studies like ours aid in identifying the variations in the circulating influenza virus strains, as well as spot emerging variants, thus abetting a vigilant control over the disease.Biography
Devanshi Gohil is a Research Scholar currently pursuing Doctorate of Philosophy in Medical Microbiology on “Molecular characterization of 2009 H1N1 pandemic influenza & seasonal influenza viruses from patients in Mumbai” at Haffkine Institute for Training, Research and Testing, under Maharashtra University of Health Sciences, Nasik, India. She has done her Post-Graduation studies in Microbiology from University of Mumbai, India. Presently, she is working as Project Coordinator on National Project entitled Laboratory Based Influenza Surveillance Plan India: Under Integrated Disease Surveillance Project (IDSP) Avian Influenza Lab Network, run by National Center for Disease Control, Government of India, New Delhi, India. She has 7 publications to her credit in national and international journals.
Email: devanshi.gohil@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi