Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
Google Scholar citation report
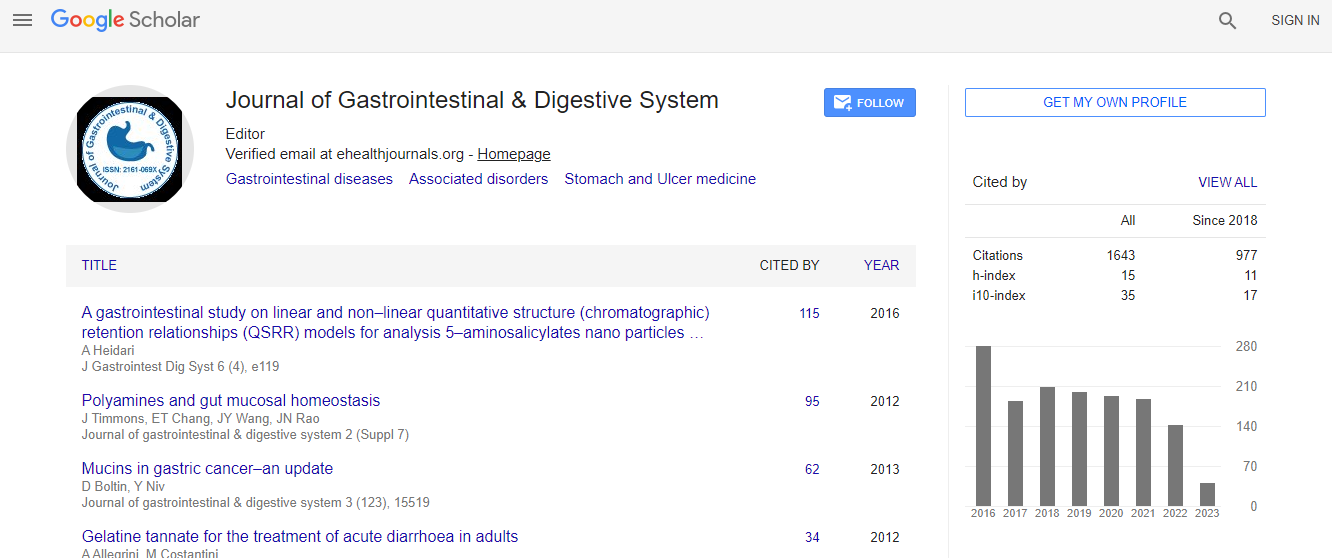
Citations : 2091
Journal of Gastrointestinal & Digestive System received 2091 citations as per Google Scholar report
Journal of Gastrointestinal & Digestive System peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Gene set-level meta-analysis of intrahepatic cholangiocarcinoma
International Conference on Gastroenterology and Liver
Manisha Mandal
MGM Medical College, India
ScientificTracks Abstracts: J Gastrointest Dig Syst
Abstract
Statement of the Problem: Intrahepatic cholangiocarcinoma (ICC) is the second most common liver cancer constituting 10% of all cholangiocarcinomas. ICC is fast growing treatment-refractory disease with poor prognosis. However, genetic mechanisms underlying ICC are still few and often limited to select genes. Therefore, the current study was carried out to gain detailed insight into the transcriptomic profile of ICC by RNA-Seq technology to identify new therapeutic opportunities. Methodology: For gene expression meta-analysis of ICC, the GEO RNA-seq data sets were fetched from GREIN (http:// www.ilincs.org/apps/grein/). A total of 3 datasets were selected to retrieve the associated metadata. The raw data counts were subjected to downstream statistical analysis (https://www.expressanalyst.ca/gene) for gene annotation, exclusion of features with > 50% missing values, and estimating them using feature-wise KNN method. A 25% filtering of both variance and relative abundance were applied followed by Log2 scale normalization. Limma was applied to compare ICC with normal cases, with p value <0.005. ComBat program was applied for adjusting data for batch effects. The validation of different datasets was achieved using Cochran├ó┬?┬?s Q tests to incorporate heterogeneities across the study model. Findings: A total of 101 samples with 18435 matched features were compared between ICC and normal cases which yielded 173 (57 up and 116 down regulated) highly significant DEGs (p value=1.0E-16). A total of 16 KEGG pathways (p<0.05) were enriched related to caffeine metabolism (CYP1A2, NAT2, XDH), steroid biosynthesis (CYP1A2, CYP7A1, CYP1A1, SRD5A2, UGT1A4), chemical carcinogenesis (CYP1A2, NAT2, CYP1A1, UGT1A4, GSTA1), metabolic pathways, bile secretion (CYP7A1, SLC2A1, ABCC4, SLCO1B3), metabolism of drugs and xenobiotics by cytochrome P450, central carbon metabolism in cancer, etc. Liver-Type Glutaminase GLS2 and PKM (Pyruvate Kinase M1/2) were the most significantly up regulated and downregulated genes with fold change values 5.0858 and -5.584 respectively. Conclusion & Significance: Gene expression meta-analyses help in the identification of molecular signatures and functional enrichment correlated with phenotypic differences between ICC and normal individuals.Biography
Manisha Mandal has her expertise in the field of molecular epidemiology of infectious and non-infectious diseases, data analysis using bioinformatic approaches towards drug development, disease modelling, next generation sequencing. She has published more than 70 research articles in her research field in different journals, one book, and presented several papers in different conferences.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi