Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
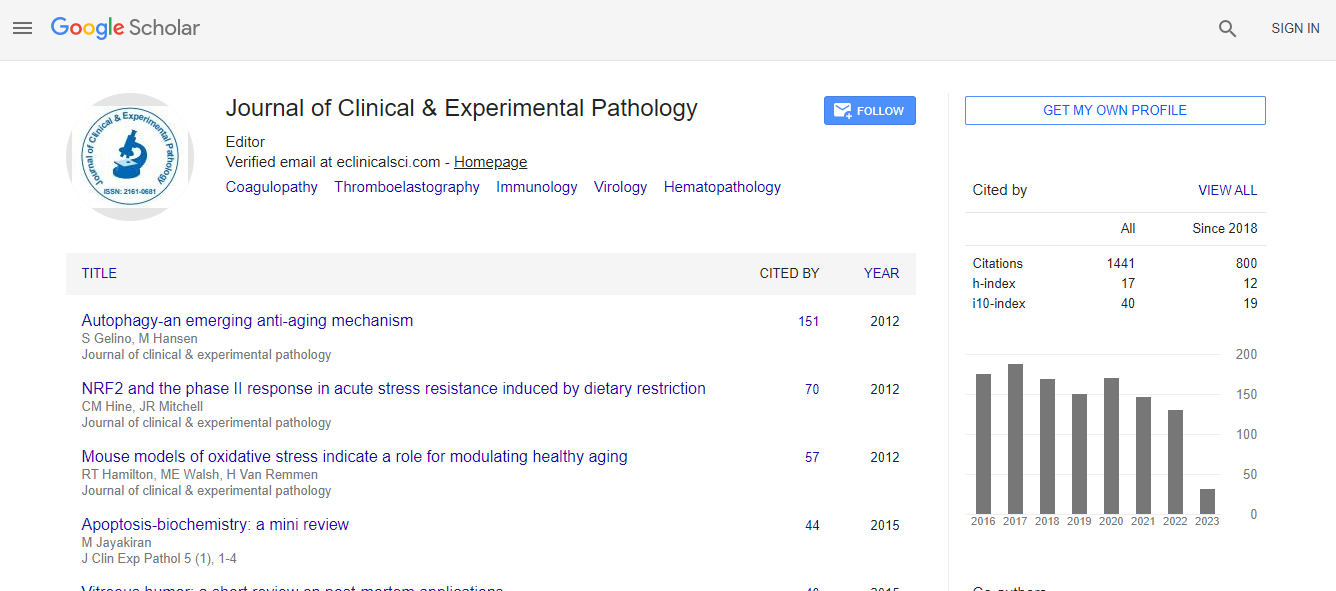
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Establishment of the recurrence prediction model of colorectal cancer using nCounter analysis system
7th World Congress on Molecular Pathology
Xiaohan Shen
Clinical Pathology Diagnosis Center, China
Posters & Accepted Abstracts: J Clin Exp Pathol
Abstract
Background: As one of the most common malignancy, colorectal cancer (CRC) poses a serious threat to human health. Both CRC incidence and mortality rates continue to increase worldwide. The establishment of recurrence prediction model of CRC has a very important practical significance. Methods: 37 gene mRNA expression levels in 473 CRC tissues were detected using nCounter analysis system. Differentially expressed genes were screened out between the recurrent group and the non-recurrent group. The recurrence prediction model of CRC was established by using the binary Logistic regression analysis. Results: A 37 gene prediction model was generated by using the binary logistic regression analysis. The univariate analysis (Log- Rank) indicated that DFS was significantly worse in the recurrent group than in the non-recurrent group both in training group and testing group (P<0.05). The univariate analysis also indicated that DEPDC1 mRNA expression were significantly higher in nonrecurrent group than those in recurrent group (P<0.05). DEPDC1 and TNM stage had significant correlation with DFS (P<0.05). The Cox proportional hazards regression indicated that DEPDC1 mRNA expression was an independent factor for CRC patients. Conclusion: In this part, we have validated the expression of differentially expressed genes and established the recurrence prediction model of CRC using nCounter analysis system which is a high-throughput platform, further study will be done to verify and optimize the prediction model in the future.Biography
Email: 12111230001@fudan.edu.cn

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi