Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
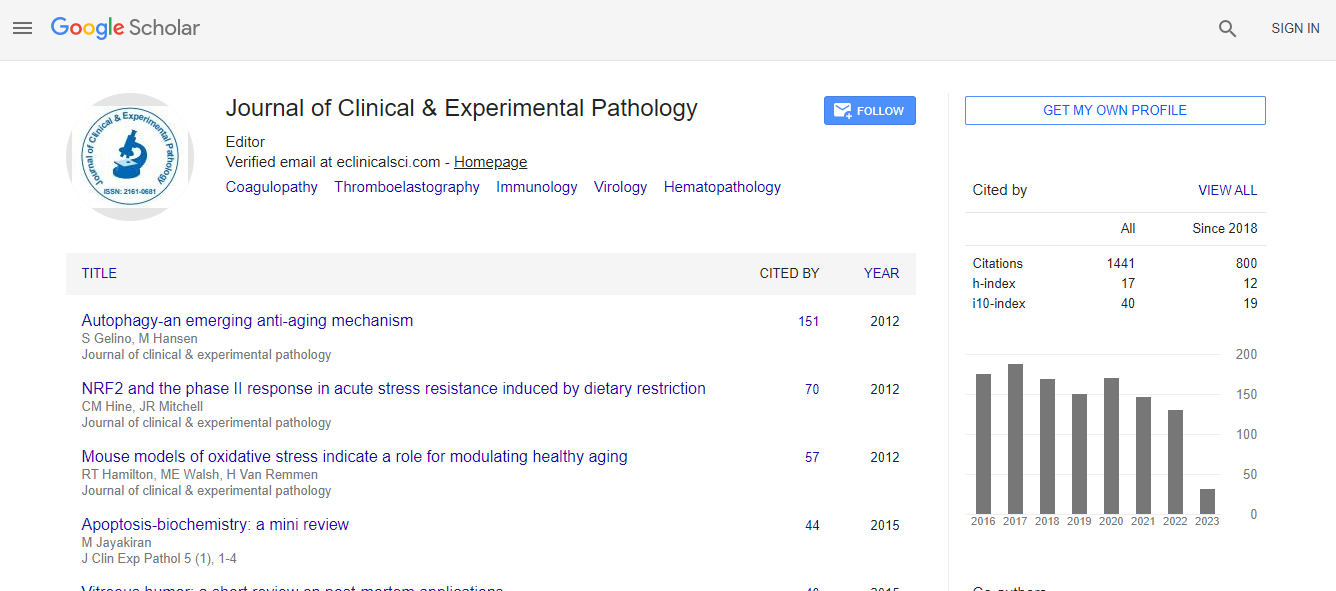
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Drug-responsive chromatin structures and the underlying genetic alterations in leukemia
13th International conference on Pathology and Molecular Diagnosis
Jason X Cheng
University of Chicago Medical Center, USA
Posters & Accepted Abstracts: J Clin Exp Pathol
Abstract
Chemical modifications of DNA/histone play an important role in organization of human chromatin into distinct structural domains that control gene expression, stem cell differentiation and tumorigenesis. Drugs that target various chromatin modifiers have become one of the promising treatments for many types of cancer including solid tumors and hematologic malignancies such as myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). However, most, if not all of the cancers treated with epigenetic drugs eventually develop drug resistance and render epigenetic drugs ineffective in cancer patients. The mechanisms underlying the selectivity and efficacy of epigenetic-modifying drugs are still unknown. Therefore, a major challenge in today├ó┬?┬?s cancer treatment is to unravel the mechanisms of drug resistance and to develop strategies to prevent or reverse drug resistance in various types of cancer. In this study, we developed a new method to simultaneously measure 5-methylcytosine (5-mC) and hydroxymethylcytosine (5-hmC). CDMIAs revealed significantly drug-responsive changes in 5-mC/5-hmC at the promoters of differentiation/lineage-controlling genes such as PU.1/SPI1. Immunoprecipitation experiments demonstrated lineage-specific, drug-sensitive interactions between the PU.1/SPI1 and GATA1 transcription factors and the DNA/histone modifying complexes. ChIP-seq and chromatin conformation capture (3C) showed that distinct chromatin structures at the gene locus in a lineage-specific manner. Importantly, novel mutations in TET2, TET3, DNMT3L and PU.1/SP1 were revealed by genome-wide sequencing and confirmed by Sanger sequencing. These mutations correlated with the altered interactions between PU.1/SPI1 and the DNA/histone modifying complexes and predicted the responses to epigenetic modifying drugs. Examination of clinical specimens from patients with MDS confirmed the presence of distinct lineage/differentiation-specific chromatin structures. These results demonstrate the importance of functional genomics in the pathogenesis of MDS and leukemia and may identify novel therapeutic targets.Biography
Email: Jason.Cheng@uchospitals.edu

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi