Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
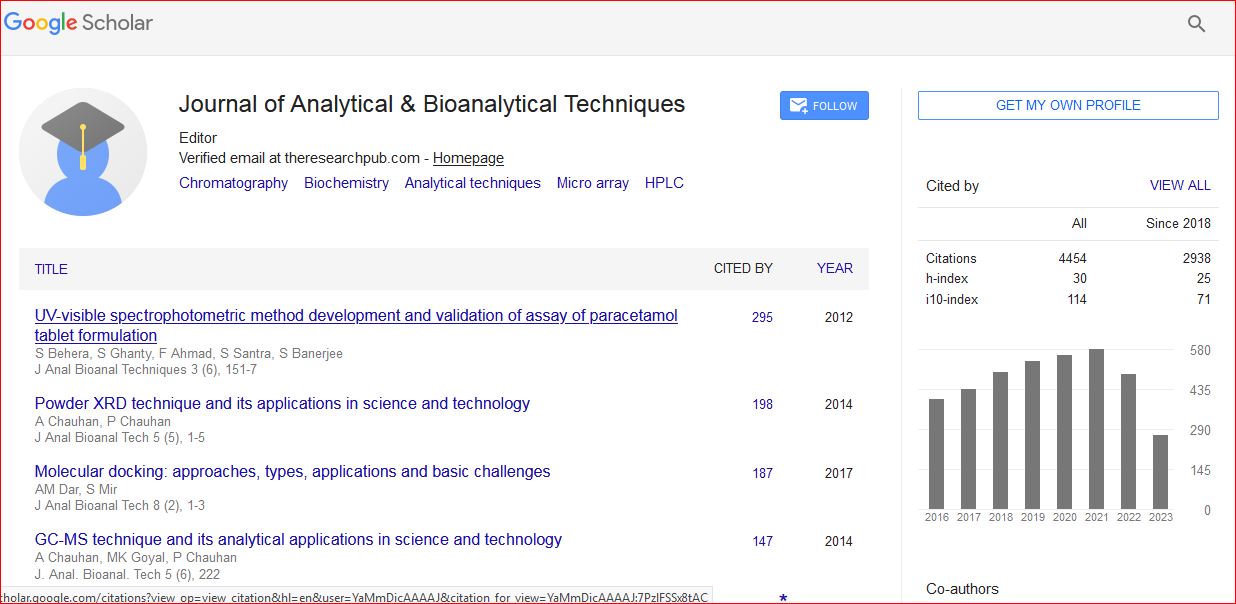
Google Scholar citation report
Citations : 6413
Journal of Analytical & Bioanalytical Techniques peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Euro Pub
- ICMJE
Useful Links
Related Subjects
Share This Page
Differential proteomics by label-free quantification for early diagnosis and prognosis of cancers
3rd International Conference and Exhibition on Analytical & Bioanalytical Techniques
Shama P. Mirza
ScientificTracks Abstracts: J Anal Bioanal Techniques
Abstract
In the last few years, differential proteomics has gained popularity due to its ability to distinguish proteome of different states by comparative analysis. This has a greater significance in identifying disease vs. healthy condition, and thereby advanced further to the application of early detection, diagnosis and prognosis of diseases using mass spectrometry (MS)-based protein quantification.Several strategies using labeling and label-free approaches have been established for both relative and absolute quantification of proteins. Recent developments in the MS instrumentation, extensive advances in bioinformatics and computing power facilitated protein quantification by label-free methods. Label-free quantification overcomes the expensive and extensive workflows required in the labeling techniques.In our laboratory, we are using a label-free quantification approach called spectral counting for the identification of disease-specific biomarkers for early diagnosis and prognosis of cancers, specifically glioblastomamultiforme (GBM) and endometrial cancer (EC). In this study, tumor biopsies and plasma/serum samples were analyzed by SDS-PAGE for minimizing the complexity of the proteome before analyzing by MS using nanoAquity UPLC-LTQ OrbitrapVelos MS. Data analysis was carried out using SEQUEST algorithm for protein identification and Visualize software for quantification of identified proteins using spectral counting method. In the GBM study, we identified 2214 � 121 proteins in tissue biopsies and 853 � 52 proteins in plasma samples, and found 883 � 71 in tumor and 363 � 56 proteins in plasma to be differentially modified (p â�?¤ 0.05). In GBM patients, 46 and 21 proteins were identified exclusively in tumors and plasma, respectively compared to controls. We further characterized two of the potential biomarkers pigment epithelium derived factor (PEDF) and brevican core protein (BCAN) using Western blotting and MS. We observed that the protein expression and possible posttranslational modifications (PTM) of these candidate biomarkers to be altered among GBM patients. Similarly, in the EC study, we identified an average of 1048 � 209 proteins in serum samples, and an average of 389 � 39 proteins with significant differential expression between pre- and post- surgery samples (p<0.05). Of these, nine proteins were absent in pre- surgery control samples but present in pre- surgery patient samples, which are enlisted to be the potential biomarkers for the early diagnosis of endometrial cancer. Using label-free quantification MS method, we identified tumor-specific proteins in patient samples, which could be the potential biomarkers for early diagnosis in EC and prognostic markers in GBM. The emerging new knowledge of protein markers and their PTMs identified from this study is expected to represent a major step forward for better understanding of the fundamental biology of cancers and for giving deeper insight into the pathogenesis of disease, thereby identifying novel therapeutic targets for optimal therapy.Biography
Shama Mirza is an Assistant Professor in biochemistry and biotechnology & bioengineering center at Medical College of Wisconsin, Milwaukee, USA. After herPhD in chemistry (mass spectrometry), shedid herpostdoctoral research in mass spectrometry-based proteomics at the University of Texas Southwestern Medical Center at Dallas and at Medical College of Wisconsin. Herresearch focuses on developing novel technologies for the comprehensive characterization of cellular proteomes using mass spectrometry in order to better understand protein functions and interactions under normal and disease states; primary focus being on glioblastomamultiforme, endometrial cancer, heart failure with preserved ejection fraction and ureteropelvic junction obstruction. Dr. Mirzais currently serving as an editorial board member for ?Proteomics Insights? and also as an invited reviewer for several other international journals in the area of biological and analytical chemistry. She has published more than 23 fully peer-reviewed papers. She is also involved in teaching PhD and medical students.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi