Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
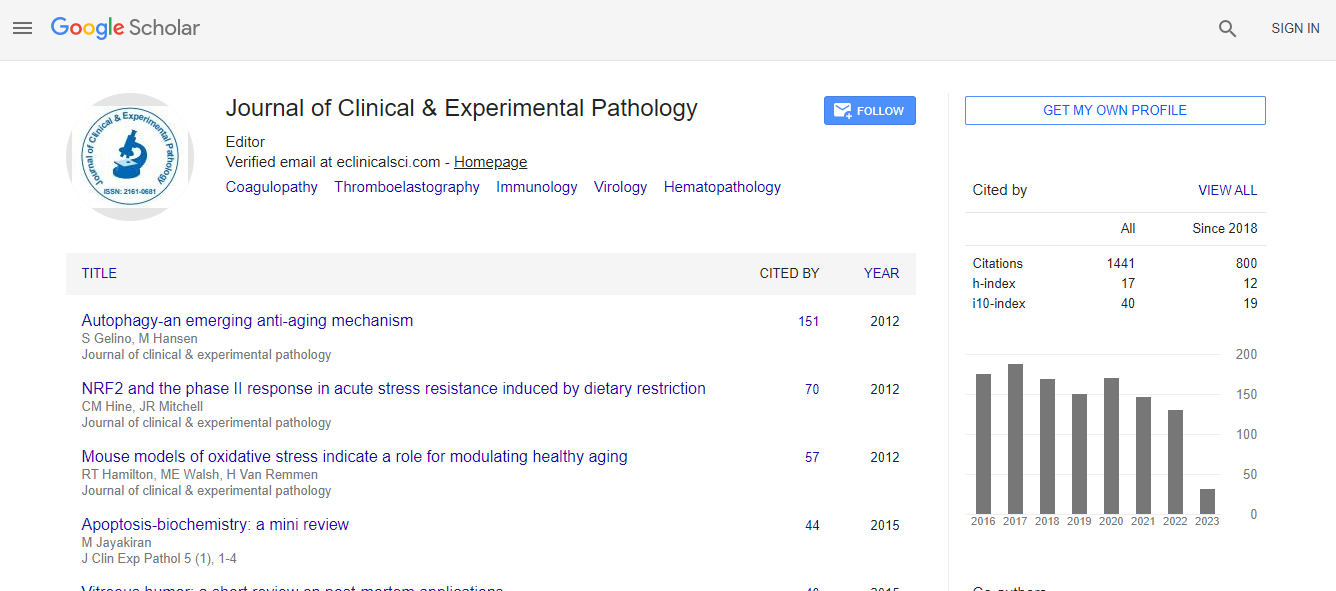
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Development of targeted re-sequencing approach for muscular dystrophy diagnosis
International Conference on Pathology
Akanchha Kesari
ScientificTracks Abstracts: J Clin Exp Pathol
Abstract
With the rapid expansion of knowledge regarding monogenic disorders, there are now often many genes associated with particular phenotypes. For example, in the muscular dystrophies, there are over 47 causative genes identified to date, with few if any phenotypic features able to distinguish between the different underlying genetic causes. Many of the muscular dystrophy genes on the panel are quite large (e.g. nebulin�147 exons;dystrophin � 79 exons; dysferlin � 55 exons). As a result, patients and physicians can often pursue an extended molecular genetics odyssey, involving send-outs to different laboratories, with commensurate high costs and long turn-around times. An alternative approach is nextgen sequencing, where all candidate muscular dystrophy genes are sequenced in parallel through either targeted re-sequencing or exome sequencing. We compared three approaches to nextgen sequencing in muscular dystrophy patients; RainDance targeted amplification of 1,841 amplimers covering the exons and exon/intron boundaries of 47 muscular dystrophy genes, and exomic sequencing (with and without a 47 gene filtering step). The RainDance unit fuses individual microbubbles of the amplimer mix with patient DNA. The emulsion is then PCR amplified, and amplimers were sequenced using 2nd(Illumina) or 3rd generation (Pacific Biosciences) sequencers. In parallel, whole exome sequencing was done on muscular dystrophy patients and was analyzed with or without the 47 gene filter. Data was assembled and analyzed using the NextGENe software and variants were called and reported accordingly.The observed advantages and disadvantages of each approach will be described.Biography
Akanchha Kesari is molecular biologist and geneticist by training; she had pursued a number of complex and interdisciplinary projects, although the focus is always on human genetic disease. Her research project has been focused to define the molecular basis for the muscular dystrophies, both with regards to identification of causative genes, and biochemical pathways immediately downstream of the primary genetic defect. Recently, she has been awarded the T32 grant in Genetics and Genomics of Muscle from NIAMS (NIH) to purse my reasearch in development of next-generation sequencing technology for muscle disease. Along with that, she is also in the fellowship training program of National Human Genome Research Institute (NHGRI) in Clinical and Molecular Genetics. Suceessful completion of the trianing will help her to become the certified molecular geneticist by American Boards of Medical Genetics (ABMG).

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi