Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
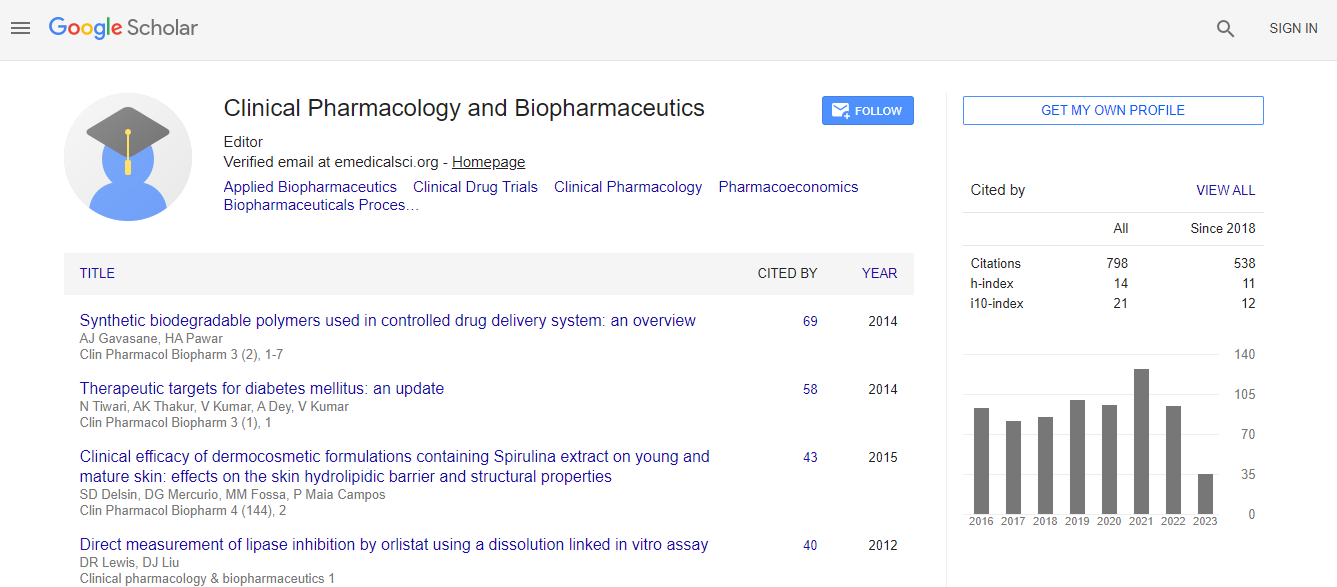
Google Scholar citation report
Citations : 1089
Clinical Pharmacology & Biopharmaceutics received 1089 citations as per Google Scholar report
Clinical Pharmacology & Biopharmaceutics peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Genamics JournalSeek
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Design, docking, synthesis and in vitro binding studies of few benzamide derivatives with glucokinase enzyme
Pharma Middle East
Vivekanand A Chatpalliwar1, Saurabh C Khadse2 and C D Upasani1
1S N J B�s S S D J College of Pharmacy, India 2R C Patel Institute of Pharmaceutical Education & Research, India
Posters-Accepted Abstracts: Clin Pharmacol Biopharm
Abstract
Glucokinase (GK) enzyme is involved in glucose utilization in liver and has also been implemented in Glucose-dependant release of insulin in the pancreatic �²-cell. Activation of glucokinase enzyme therefore, has emerged as a strategy to increase glucose utilization. The world-wide endeavours to design compounds (GKA: Glucokinase activators) that would activate the glucokinase enzyme so as to develop suitable drugs to treat Type 2 diabetes, has culminated in library of numerous compounds; benzamide derivatives have been at mainstay amongst them. The ligand-protein interaction involves ARG63 as the anchoring residue, and many more residues as TYR210, TYR215, etc. which are submerged in a cleft between two domains. Albeit, binding of the ligands with amidine backbone of ARG63 and the hydrogen-bond interactions of the free amino group on ligands with TYR215 is believed to be involved in activation of the enzyme, the free amino group had been noticed to elicit mutagenic effects. Present work describes the design, docking of benzamide derivatives that bind with ARG63, much the way similar to the standard GKA, RO-28-1675, and interact with other set of amino acid residues in much similar way, however, without the presence of free amino group. Molecular modeling studies involved, Molecular Design Suite (VLife MDS 3.5), simulated protein 1V4S, virtual structures of standard and newly designed benzamide derivatives. The structures were docked in the catalytic site of 1V4S to obtain respective docking score (in KJ/mol) and the amino acid residue involved in each interaction. Based on the study, few molecules were selected for synthesis and other molecules were outright rejected. The criteria for such selection, scheme of reactions to synthesize selected molecules, results of their in vitro binding with glucokinase enzyme are presented. The obtained results do not promise to yield better GKA than the standard compound. However, the molecules could further be subjected to rational changes in structure and re-subject to in silico studies so as to yield molecules to activate GK.Biography
Email: vchatpalliwar@yahoo.co.in

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi