Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
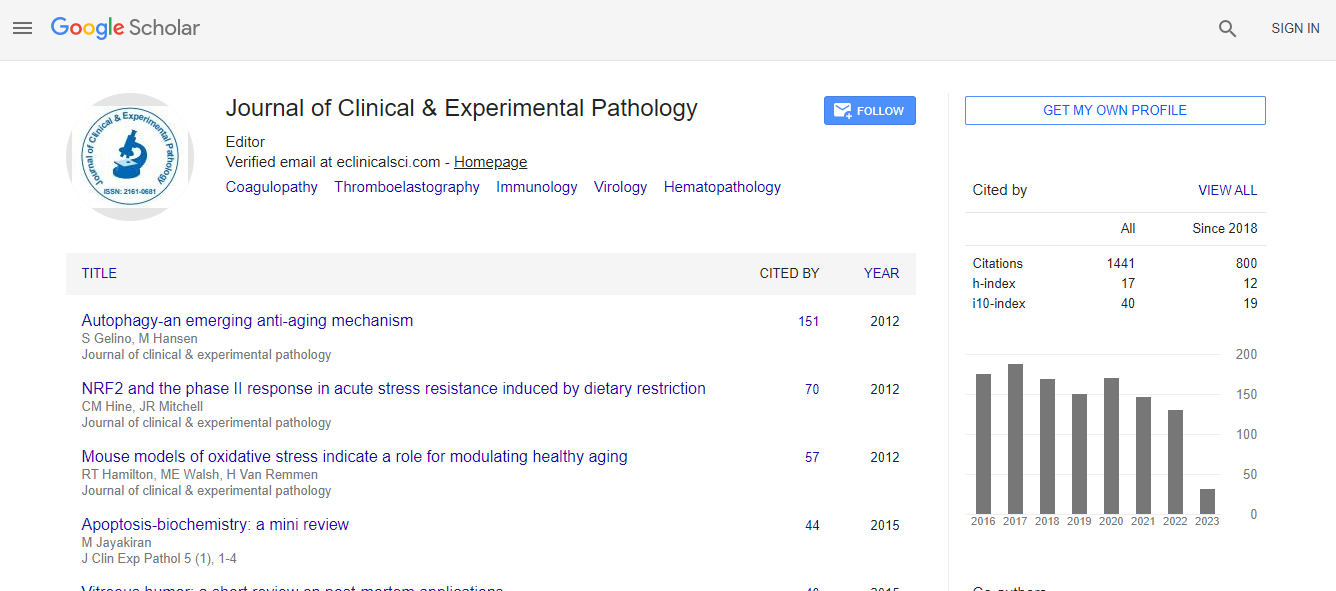
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Automated segmentation of nucleus, cytoplasm, and background of pap-smear images using a trainable pixel level classifier
Joint Event on 15th International Congress on American Pathology and Oncology Research & International Conference on Microbial Genetics and Molecular Microbiology
Wasswa William, Andrew Ware, Annabella Habinka Basaza-Ejiri and Johnes Obungoloch
Mbarara University of Science and Technology, UgandaUniversity of South Wales, UKSt. Augustine International University, Uganda
Posters & Accepted Abstracts: J Clin Exp Pathol
Abstract
Background: Cervical cancer ranks as the fourth most prevalent cancer affecting women worldwide and its early detection provides the opportunity to help save a life. Automated diagnosis and classification of cervical cancer from pap-smear images has become a necessity as it enables accurate, reliable and timely analysis of the condition’s progress. Segmentation is a fundamental aspect of enabling successful automated pap-smear image analysis. In this paper, a potent algorithm for segmentation of the pap-smear image into the nucleus, cytoplasm, and background using pixel level information is proposed. Methods: First, a number of pixels from the nuclei, cytoplasm, and background are extracted from five hundred images. Second, the selected pixels are trained using noise reduction, edge detection, and texture filters to produce a pixel level classifier. Third, the pixel level classifier is validated using test set and 5- fold cross validation using Fast Random Forest, Naïve Bayes, and J48 classification techniques. Results: An extensive evaluation of the algorithm and comparison with the benchmark ground truth measurements shows promising results. Comparison of the segmented images’ nucleus and cytoplasm parameters (nucleus area, longest diameter, roundness, perimeter and cytoplasm area, longest diameter, roundness, perimeter) with the ground truth segmented image feature parameters (nucleus area, longest diameter, roundness, perimeter and cytoplasm area, longest diameter, roundness, perimeter) yielded average errors of 0.94, 0.93, 0.02, 0.63, 0.96, 0.37, 0.13 and 0.96mm respectively. Validation of the proposed pixel level classifier with 5-fold cross-validation yielded a classification accuracy of 98.48%, 94.25% and 98.45% using Fast Random Forest, Naïve Bayes, and J48 classification methods respectively. Finally, validation with a test dataset yielded a classification accuracy of 98.48% and 98.98% using Fast Random Forest and J48 Classification methods respectively. Conclusion: This paper articulates a potent approach to the segmentation of cervical cells into the nucleus, cytoplasm, and background using pixel level information. The experimental results show that the approach gives good classification and achieves a pixel classification average accuracy of 98%. The method serves as a basis for first level segmentation of pap-smear images for diagnosis and classification of cervical cancer from pap-smear images using nucleus and cytoplasm pixel level information.Biography
E-mail: wwasswa@must.ac.ug

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi