Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
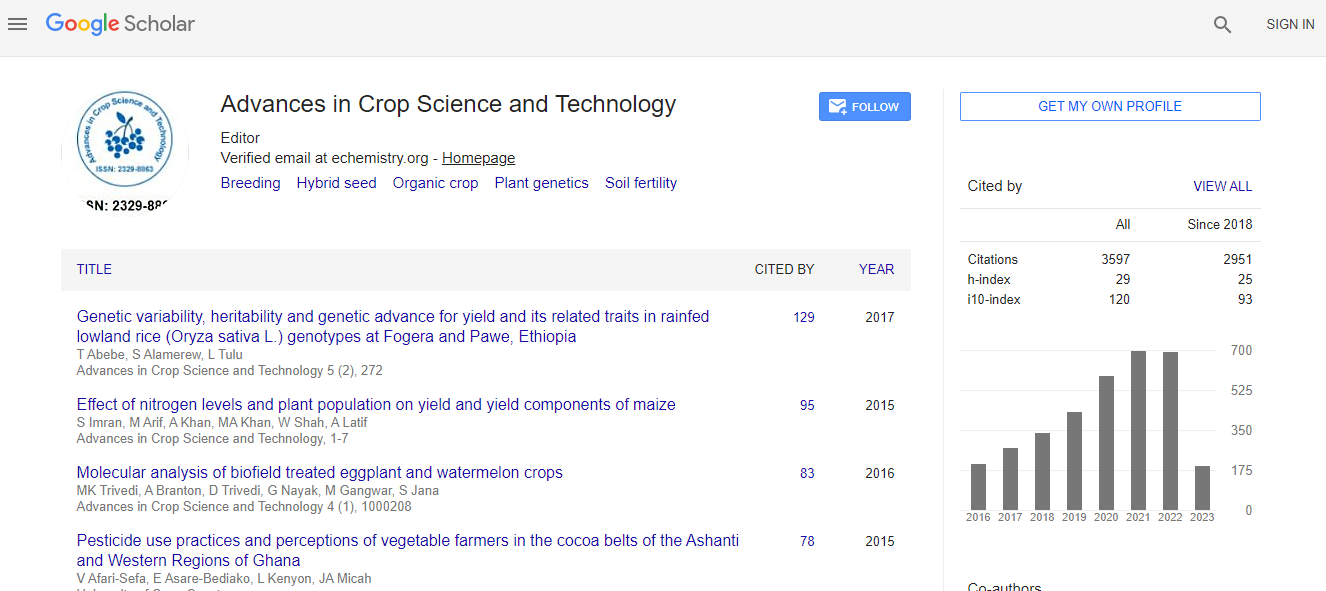
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
Assessment of candidate DNA Barcoding loci for phylogenetic relationships among Triticum species
11th World Congress on Plant Biotechnology and Agriculture
Lee Gi-An, Jung-Ro Lee, Gyu-Taek Cho, Kyung-Ho Ma and Sebastin Raveendar
National Agro-Biodiversity Center, Republic of Korea
Posters & Accepted Abstracts: Adv Crop Sci Tech
Abstract
Statement of the Problem: DNA Barcoding is widely used for identification of species based on standard DNA regions. The Consortium for the Barcode of Life (CBOL) plant-working group recommended different nuclear and chloroplast loci as the standard plant barcode. Though many loci have been proposed as a DNA barcodes in plants, standardizing regions as a DNA barcode poses a challenge in many plant families. The evolutions of the chloroplast regions combine with nuclear gens are sufficiently rapid to allow discrimination between closely related species. Material & Methods: In this study, we tested the phylogenetic utility of the DNA barcoding loci (ITS2, matK, psbA-trnH, rbcL and trnT) for efficient discrimination of species. For the assessment of the barcoding efficiency to resolve the species discrimination, a total of 113 accessions representing 18 species/sub-species within Triticum genus have been sampled. Consensus sequences of each region were manually edited with MEGA6 and the sequences were aligned and assessed for candidate DNA Barcode for phylogenetic relationships among Triticum species. Findings: Topologies of the phylogenetic trees based on combination of DNA barcode analyses were similar but a few Triticum species were placed into distant phylogenetic groups. The 113 accessions analyzed in this study were placed into three groups supported by high bootstrap values. However, the barcoding analyses were not able to discriminate some closely related Triticum species. Conclusion & Significance: We have proposed concatenated data approach to increase resolving power of candidate barcoding loci as an additional tool for phylogenetic analysis in Triticum species. However, molecular studies with more diverse markers and species will be required to clarify the ambiguities surrounding the phylogeny of these important genera.Biography
Lee Gi-An has his expertise in evaluation and characterization of plant genetic resources. He has worked in National Agro-Biodiversity Center (NAc, RDA, Republic of Korea) for more 12 years as a Researcher.
Email:gkntl1@korea.kr

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi