Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
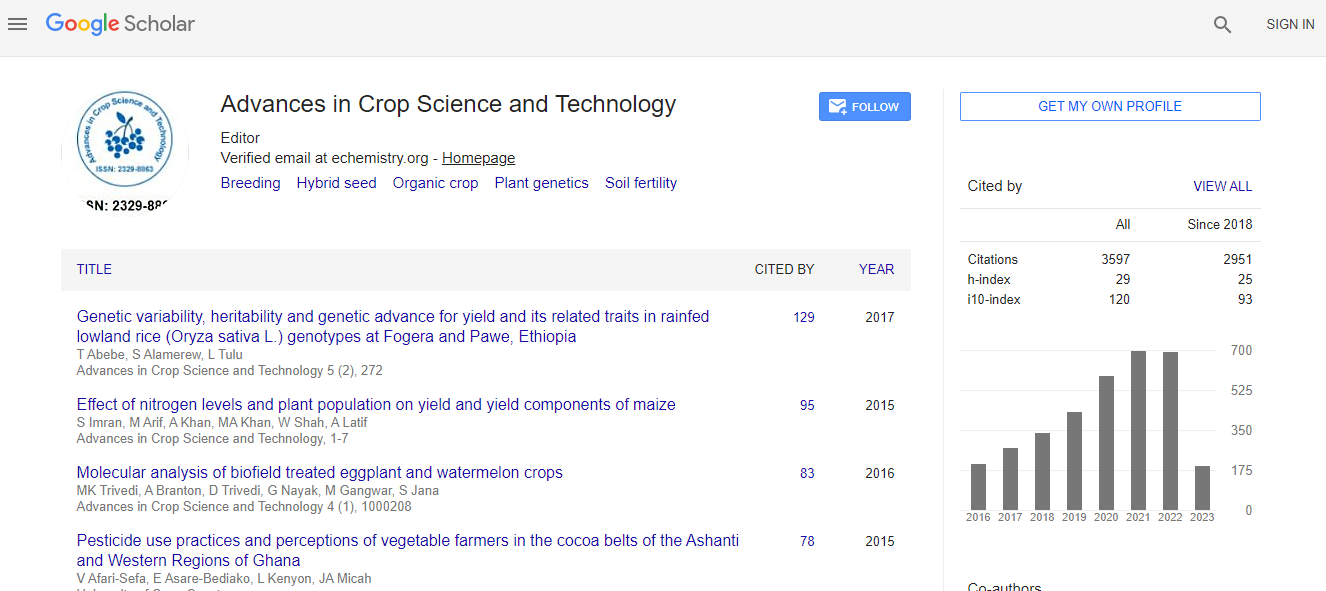
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
Application of next-generation sequencing, genome sequencing and whole genome re-sequencing to practical plant breeding: Case studies on lupin
4th International Conference on Plant Genomics
Huaan Yang
Department of Agriculture and Food Western Australia, Australia
ScientificTracks Abstracts: Adv Crop Sci Tech
Abstract
Next-generation sequencing (NGS) and whole genome sequencing bring us a deluge of genome and genetic variations. In lupin, we are applying NGS and genome sequencing technologies to help practical plant breeding through four avenues. (1) NGS as DNA fingerprinting for rapid trait-marker association discovery. Markers closely linked to disease resistance genes were successfully discovered by NGS-based RAD sequencing and were applied to marker assisted selection (MAS) in lupin. (2) Genome sequencing for developing functional markers. We completed the draft genome sequence of lupin; a diagnostic marker linked to anthracnose disease resistance gene was developed by gene annotation and further confirmed by genetic mapping. (3) Genome sequencing to developed cost-effective markers. In the last 15 years, DAFWA has developed over a dozen of gel-based InDel markers linked to various genes of agronomic traits of interest in lupin. Using the draft lupin genome sequence as templates, all previously developed InDel markers were converted into a cost-effective SNP markers to suit modern SNP genotyping platform. (4) Whole genome re-sequencing for developing diagnostic markers for MAS. We recently resequenced the whole genomes of17 lupin lines; several million markers were documented and 207,887 markers were anchored on the lupin genetic linkage map. We demonstrated two protocols of using wholegenome resequencing data for rapid development of diagnostic markers for MAS. In conclusion, the cases in lupin represent one of a few successful cases where NGS and genomes sequencing have been used as routine tools for molecular plant.Biography
Huaan Yang is a Plant Pathologist by training with BSc in 1982 and MSc in 1985. He has completed his PhD from University of Western Australia in 1992. He became a Molecular Geneticist following his development of a DNA fingerprinting technology called “MFLP” in 2001. His lab has been the only lab for large-scale application molecular markers on MAS in plant breeding on legume grain crops in Australia since 2004. Since 2011, his lab has been fully switched into NGS and genome sequencing based approaches for pre-breeding in lupin.
E-mail: huaan.yang@agric.wa.gov.au

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi