Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
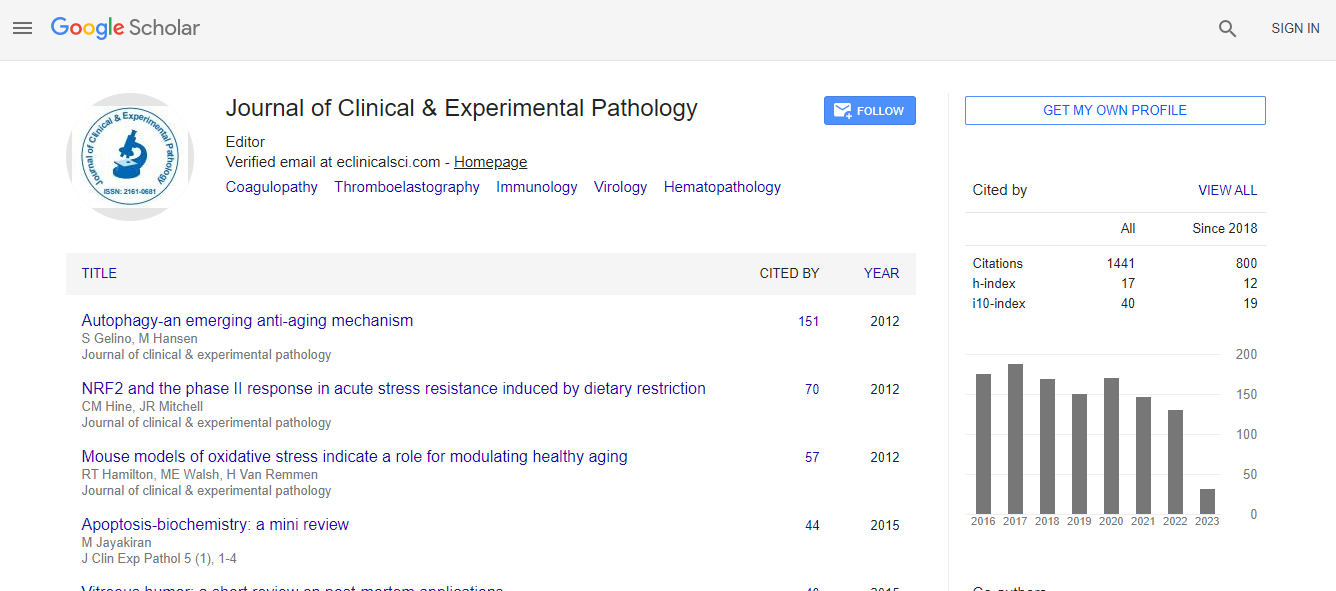
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
A framework for translating advances in molecular genetic technologies into clinical laboratory practice
13th International conference on Pathology and Molecular Diagnosis
Malgorzata Jaremko and Alexander Bisignano
Phosphorus Diagnostics, USA
ScientificTracks Abstracts: J Clin Exp Pathol
Abstract
Advances of the molecular diagnostic testing platforms, including development and implementation of NGS based genetic testing contribute to the improvement of disease prediction, diagnosis, and treatment. However, the future of genomic medicine relies on the capability of molecular genetics laboratories to develop and validate evidence-based and costeffective laboratory tests. These laboratories face many challenges including establishing clinical utility, validating analytical performance of laboratory developed tests, and managing costs of platform development and subsequent consumables. Along with the molecular and instrumentation challenges, laboratories are faced with a myriad of software options (e.g., Galaxy, Amazon, GATK, BaseSpace, and Clarity) when establishing a reliable bioinformatics pipeline and LIS system. Finally, there is a lack of consensus and consistency in the quality standards across the industry (e.g., read depth, variant curation, and clinical validation structure). In this study, we present a framework for the consistent development of accurate, high-quality, NGS diagnostic tests. Our process is broken into stages from gene selection through clinical validation and implementation. Based on the experience in our own CLIA-laboratory, we present lessons learned in the development of NGS targeted panels for sequencing and CNV analysis for various indications including infertility, hereditary cancers, arrhythmias, cardiomyopathies and lipidemias.Biography
Malgorzata Jaremko has completed her PhD in Pharmacogenomics from Wroclaw Medical University and Postdoctoral Clinical Fellowship from Mount Sinai School of Medicine, NY. She is board certified by American Board of Medical Genetics and Genomics in Clinical Molecular and Clinical Biochemical Genetics; and she is Fellow of American College of Medical Genetics and Genomics, as well as National Academy of Biochemistry. She has extensive experience in directing clinical molecular laboratories, and currently serves as the Senior Director, Clinical Laboratory & Molecular Diagnostics, and CLIA-Director of Phosphorus Diagnostics genetic testing laboratory.
Email: m.jaremko@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi