Research Article Open Access
Yeast Extract as the Most Preferable Substrate for Optimized Biosurfactant Production by rhlB Gene Positive Pseudomonas putida SOL-10 Isolate
| Muneer Ahmed Qazi1, Zulfiqar Ali Malik1,2, Ghazi Dino Qureshi1, Abdul Hameed1 and Safia Ahmed1* | |
| 1Department of Microbiology, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad 45320, Pakistan | |
| 2Department of Microbiology, Faculty of Natural Sciences, Shah Abdul Latif University, 66111 Khairpur Mir’s, Sindh-Pakistan | |
| Corresponding Author : | Safia Ahmed Department of Microbiology Faculty of Biological Sciences Quaid-i-Azam University Islamabad 45320, Pakistan Tel: +9251-90643009 E-mail: safiamrl@yahoo.com |
| Received: June 09, 2013; Accepted: September 21, 2013; Published: September 27, 2013 | |
| Citation: Qazi MA, Malik ZA, Qureshi GD, Hameed A, Ahmed S (2013) Yeast Extract as the Most Preferable Substrate for Optimized Biosurfactant Production by rhlB Gene Positive Pseudomonas putida SOL-10 Isolate. J Bioremed Biodeg 4:204. doi:10.4172/2155-6199.1000204 | |
| Copyright: © 2013 Qazi MA, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Oil contaminated sites are enriched source of microorganisms that produce a variety of surface active amphiphilic compounds known as biosurfactants. Pseudomonas putida SOL-10 strain isolated from oil contaminated soil of Fimkassar oil field, Chakwal, Pakistan, was identified by standard morphological, biochemical and 16S rRNA sequence analysis methods. SOL-10 strain was initially screened for biosurfactant production using oil spreading test and then manifestation of rhlB (rhamnolipid) gene was confirmed by PCR using gene-specific primers. Maximum biosurfactant production in terms of surface tension (29.9 mN m-1) and emulsification index (E24, 73.45%), was achieved when the strain was grown in MSM supplemented with yeast extract (1.5-2 %, w/v) and urea (0.1 %, w/v) as carbon and nitrogen sources, respectively, and the physical parameters were adjusted at pH 7.0, temperature 30°C, 150 rpm agitation speed. The biosurfactant emulsified various hydrocarbons tested, being more effective against xylene and kerosene (85.19% and 70.59%, respectively). The crude biosurfactant also showed stability at a wide range of temperature (25-80°C), pH (1-9) and salt concentration (1-5%, w/v). The stability and hydrocarbon emulsifying potential of the biosurfactant indicated its possible use as decent contender for future environmental applications like biodegradation and bioremediation of organic pollutants.
| Keywords |
| Yeast extract; rhlB gene; Pseudomonas putida; biosurfactant; Hydrocarbon emulsification |
| Introduction |
| Biosurfactants are surface active amphiphilic compounds produced mainly by various microorganisms [1]. Generally, surface active compounds are of two types, i.e. chemically synthesized (surfactants) and biologically synthesized (biosurfactants). The chemical surfactants are produced from petrochemicals and are considered to be environmentally unsafe because they can lead to an ecological imbalance due to toxicity and less biodegradability [2,3]. On the contrary, biosurfactants are environment friendly because these are less toxic and biodegradable. The other advantages of the biosurfactants over chemical surfactants include their specific activity even at extreme environmental conditions and production from renewable resources [4-6]. The biosurfactants have widely been applied in various fields like food and agriculture, detergents and cosmetics, environmental cleanup and oil recovery, biomedical and therapeutics [6-8] during recent years. |
| Due to imminent petroleum crisis worldwide, the current market demand for industrially sustainable biosurfactants has grabbed the attention of many companies for commercial production of costeffective and environment friendly substitutes to synthetic surfactants [3,9,10]. Therefore, the global market of biosurfactants, growing at a CAGR of 3.5%, is expected to reach USD 2,210.5 million in 2018 [11]. This in turn, has also encouraged a tremendous progress in the field of biosurfactant research and has attracted the attention of many researchers towards relatively unexplored natural resources. There is a constant search for new and novel microbial strains having capability for the production of value added green chemicals, new energy sources and new drugs of therapeutic potential. |
| A large variety of surface active compounds have been isolated from hundreds of microorganisms including glycolipids, phospholipids, lipopeptides, lipoproteins, glycoproteins, polymeric and particulate structures etc. [10,12]. Among microorganisms, Pseudomonas aeruginosa for rhamnolipids, Bacillus subtilis for surfactin and Candida spp. for sophorolipids and mannosylerythritol lipids are most extensively studied [1,13]. The largest group of rhamnolipid producing bacteria belongs to the genus Pseudomonas [14]. Apart from a constant debate among the scientists about the prevalence of rhamnoilipd genes in bacteria other than Pseudomonas species, some novel strains of bacteria having unique ability to produce rhamnolipidshave recentlybeen reported [15,16]. Rhamnolipid production in few pathogens including Burkholderia mallei, B. pseudomallei and the nonpathogenic B. thailandensis, also been reported [17]. In bacteria, the primary biosynthetic and regulatory genes are present in an rhlABRI gene cluster which is involved in the production of rhamnolipids [18]. There’s a continuous debate among the scientific community over prevalence of rhamnolipid genes in bacteria other than P. aeruginosa. In the current study, a bacterial strain Pseudomonas putida, isolated from one of the oil fields of Pakistan, was screened and optimized for rhamnolipid production. P. putida, considered to be a nonpathogenic member of Pseudomonads, is found mostly in moist environments like soil and water (especially in rhizosphere). It is found to be involved in plant protection from pests, plant growth promotion, and environmental clean-up of persistent organic pollutants. The bacterium has also been considered to have enormous potential for biotechnological applications [19]. |
| Although, biosurfactants are advantageous over chemical surfactants available in the market, the limited yields, foaming issues, poor recoveries and higher production- as well as purification-costs are still major hurdles in their production at commercial scale. Apart from that, all microbial processes are influenced by environmental and culture conditions that also play a major role in type and productivity of the biosurfactants [4,20]. Therefore, the major goal of this study was to evaluate the effect of cultural and environmental conditions on optimal biosurfactant production by P. putida SOL-10 isolate. |
| Materials and Methods |
| Microorganism and culture maintenance |
| The bacterial isolate SOL-10, isolated from Fimkessar oil field, Chakwal, Pakistan was obtained from culture collection of Microbiology Research Labs., Quaid-i-Azam University, Islamabad. The strain was already known as a potential crude oil-biodegrading agent [21]. The bacterium was routinely cultured on nutrient agar and preserved on nutrient agar slants at 4°C in refrigerator for routine use. Culture stocks of pure strain were prepared in nutrient broth with glycerol (20%) for long term preservation at -70°C. The isolate SOL-10 was screened for biosurfactant producing ability using agar plate-based oil spreading test [22]. |
| Physiological and biochemical characterization of SOL-10 isolate |
| The bacterial strain was primarily characterized on the basis of Gram reaction, motility, spore and capsule formation. The physiological and biochemical characterization of SOL-10 strain that included: hydrolysis of starch, lipids and casein; catalase, oxidase, indole production citrate and carbohydrate (lactose) utilization tests, was performed as per standard methods according to Bergey’s Manual of Determinative Bacteriology [23]. The Analytical Profile Index (API) was also obtained to further confirm biochemical characteristics of SOL-10 strain with API 20E Kit and API identification software using online APIweb services (BioMérieux, France). |
| 16S rRNA based identification |
| The bacterial genomic DNA was extracted from overnight pure culture of SOL-10 strain using ethanol precipitation method [24]. Briefly, 5 to 6 pure bacterial colonies were picked and suspended in 240 μL distilled water. To this, 20-30 μL of sodium dodecylsulfate (SDS), 80 μL of PK(Proteinase K) buffer and 40 μL of PK were mixed and incubated at 55°C for 1 h for disintegration of cells. 100 μL of 6M NaCl was added. Centrifugation was done at 14,000 rpm for 5 min. Supernatant was taken and 1 mL of chilled 100% ethanol was added in order to precipitate DNA. Tubes were then mixed well, centrifuged at 12,000 rpm and supernatant was discarded. Washing of the pelleted DNA was carried out twice with 1 mL of 70% ethanol, centrifuged and supernatant was discarded. The pellet was air dried and 100 μL of TE buffer was added. The partial sequencing of the 16S rRNA gene was commercially carried out at the Genomic Division, Macrogen Inc., Seoul, Korea, using universal amplification and sequencing primers (Table 1) and ABI PRISM Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit (PE Biosystem, USA). Electrophoresis of sequencing reaction was completed using the automated ABI PRISM 3730×l DNA Sequencer (Applied Biosystems, USA). |
| Phylogenetic correlation analysis |
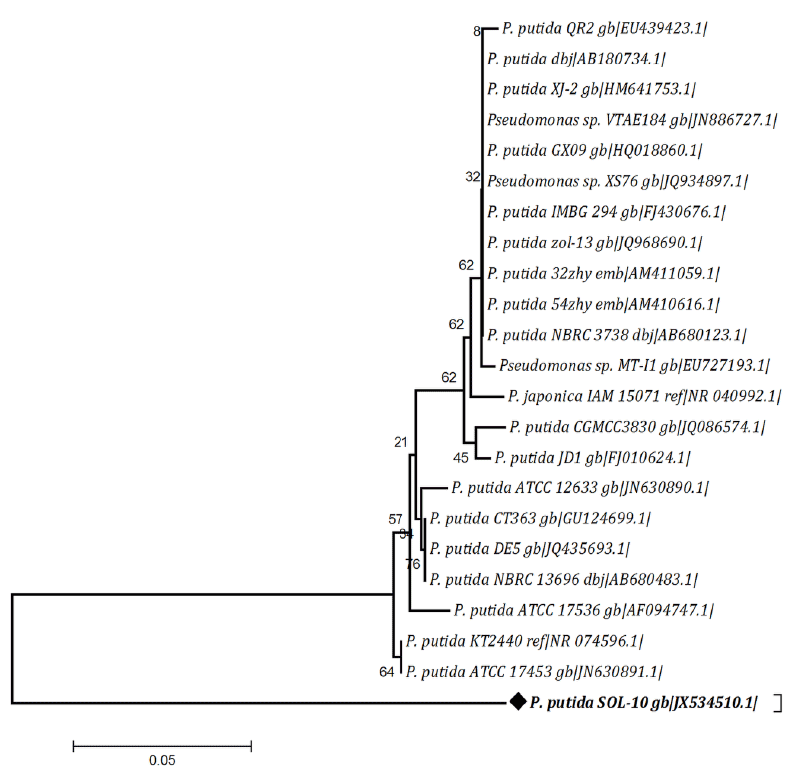
| The partial sequence of the 16S rRNA gene obtained in this study was analyzed and compared with nucleotide sequence databases in the National Center for Biotechnology Information (NCBI) website using Basic Local Alignment Search Tool (BLAST) program (http://www. ncbi.nlm.nih.gov/BLAST), in order to confer percentage sequence similarities. The evolutionary history of SOL-10 strain was inferred using the Neighbor-Joining (NJ) method [25]. The evolutionary distances were computed using the Maximum Composite Likelihood (MCL) method [26]. Phylogenetic analyses were conducted in Molecular Evolutionary Genetics Analysis (MEGA) software (Version 4.0) [27]. |
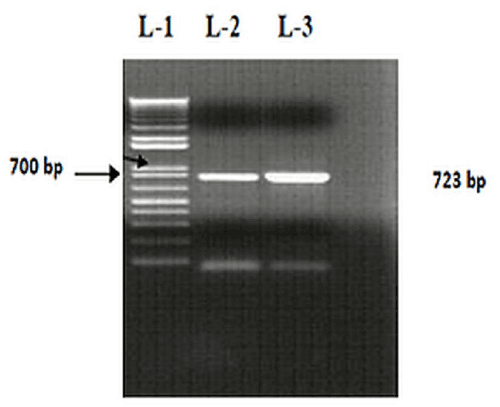
| Molecular screening for rhlB gene |
| PCR for rhlB gene amplification was carried out was in Thermocycler 2700 (Applied Biosystems, USA) using gene-specific primers (kpd-F 5’-GCCCACGACCAGTTCGAC-3’ and kpd-R 5’-CATCCCCCTCCCTATGAC-3’) according to Danashekar et al. [28]. The PCR amplification cycle consisted of an initial denaturation step at 94°C for 5 min, followed by 34 cycles of 25 sec at 94°C and annealing at 54°C for 40 sec. Afterwards, an elongation cycle (72°C for 50 sec) was followed by a final extension step (72°C for 6 min). The final PCR product (10 μL) was resolved with gel electrophoresis using 1 % agarose gel in 0.5× TBE buffer against 1 kb DNA marker (Fermentas) and ethidum bromide-stained bands were visualized under UV-transilluminator. The gel images were taken with Bio-Rad gel documentation system (Bio-Rad Laboratories, Inc., USA). |
| Culture conditions for growth and biosurfactant production |
| Mineral salts medium (MSM) as described by Abouseoud et al. [29], with slight modification, was used throughout the fermentation experiments. The medium contained (g L-1 of deionized water): Na2HPO4, 2.2; KH2PO4, 1.4; MgSO4.7H2O, 0.6; FeSO4.7H2O, 0.01; NaCl, 0.05; CaCl2, 0.02; and 0.1 mL of trace elements solution containing g L-1 of deionized water: ZnSO4.7H2O, 2.32; MnSO4.4H2O, 1.78; H3BO3, 0.56; CuSO4.5H2O, 1.0; NH4MoO4.2H2O, 0.39; KI, 0.66.The pH of medium was adjusted to 7.0 0.2 using 1 M HCl and 1M NaOH and was autoclaved at 121°C and 15 lb pressure for 20 min. The carbon and nitrogen sources were separately sterilized and added to the production medium at 2% and 0.1% concentrations respectively. All the chemicals were purchased from Sigma (Sigma-Aldrich, USA) |
| Preparation of the inoculum |
| A 5% of the inoculum, 16-18 h grown culture in nutrient broth was separated by centrifugation at 5,000rpm and washed twice with saline. After second centrifugation, the biomass pellet was aseptically re-suspended into MSM and maintained with an optical density of 1.0. This cell suspension was then used as inoculum in production medium at 5% (v/v) concentration. |
| Optimization of cultural conditions for biosurfactant production |
| Different environmental as well as nutritional factors were optimized for the production of biosurfactant and the cultivation was performed in 250 mL shake flasks within shaking incubator at certain range of pH (2.0 to 10.0), temperature (25, 30, 37 and 45°C)and agitation speed (0, 120, 150 and 200 rpm). However, the effect of nutritional factors like carbon source (yeast extract, glucose, glycerol, sucrose, η-hexadecane, xylene, kerosene and olive oil) and nitrogen source (KNO3, NH4NO3, NaNO2, NaNO3, urea, yeast extract, vitamin B2 (Riboflavin), L-Leucin and L-Arginine) on the biosurfactant production by SOL-10 strain was evaluated. For that purpose, shake flask experiments were performed in 250 mL flasks containing 100 mL MSM plus 2% and 0.1% of each carbon and nitrogen source, respectively, at 30°C and 150 rpm for 5 days. In another experiment, cultivation was also performed with different concentrations of the most suitable carbon source at 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0 and 4.5% (w/v), in order to determine the effect of varying carbon source concentration on biosurfactants producing ability of the isolatein order to optimize the yield. |
| Analytical methods |
| The qualitative and quantitative analysis of biosurfactantscontaining cell-free supernatant (CFS) of culture broth was performed. The samples were collected after every 24 h, centrifuged at 12,000 rpm for 15 min at 4°C prior to analysis. All the analyses were performed in triplicates and the averages of the three were taken. The Oil Displacement Activity (ODA) test was performed according to Rodrigues et al. [30]. The CFS (20 μL) was dropped on crude oil-film over surface of 40 mL distilled water in 90 mm petri dish. The zone of oil repellence was visualized and measured in millimeter (mm), quickly after performing the assay. The percentage emulsification index (E24) of the CFS and crude extract was determined according to the method of Cooper and Goldenberg using Eq. 1 [31]. |
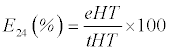 (1) (1) |
| Where ‘eHT’ denotes the emulsion height and ‘tHT’ indicates total height of the solution. The heights were measured in terms of centimeters (cm) in triplicates and the means were calculated. |
| The surface tension (SFT) of the cell-free broth, and crude biosurfactant solution was measured with a digital tensiometer (EasyDyne K20, KRUSS GmbH, Germany) using standard Whilmay plate method at room temperature according to the manufacturer’s instructions. All the measurements were taken as the mean of five measurements in mN m-1 ± SD. |
| Isolation and stability of crude biosurfactant |
| The 72 h old culture broth was centrifuged at 12,000 rpm (15 min, 4°C) and filtered (0.45 μm syringe filters). The cell free supernatant (CFS) was acidified with 6N HCl to pH 2 and left overnight at 4°C. After centrifugation, the precipitates were collected and re-dissolved in phosphate buffer (pH 7). The biosurfactants were then extracted three times with equal volume of chloroform-methanol (2:1) and pooled. The solvent was evaporated by using rotary evaporator at 45°C. The remaining brown extract served as crude biosurfactant. |
| The effect of a wide range of pH (1-11), temperature (25-121°C), and salt concentrations (1-7%) on activity and stability of the crude biosurfactant was tested. Briefly, the crude biosurfactant (100 mg/50 ml, w/v) was dissolved in range of pH buffers (1, 3, 5, 7, 9, 11) and incubated for 1 h. Subsequently, the crude biosurfactant (100 mg/50 ml, w/v), dissolved in most suitable buffer, was exposed to different temperature treatments (at 25, 40, 60, 80, 90, 100°C) in water bath for 1 h and also autoclaved at 121°C under 15 lb pressure. Different concentrations of NaCl (1, 2, 3, 4, 5, 6 and 7%) were added to the crude biosurfactant solution (100 mg/50 ml, w/v) prepared in distilled H2O and incubated for 1 h at room temperature. The activity and stability of the biosurfactant was determined after each treatment in terms of surface tension, emulsification index and oil displacement activity. |
| Hydrocarbon emulsifying potential of biosurfactant |
| The hydrocarbon emulsifying capability of the biosurfactant against different hydrophobic sources was evaluated. For that purpose, equal volumes of each hydrocarbon (i.e., n-hexane, n-hexadecane, kerosene, xylene, glycerol and olive oil) and CFS were mixed at high speed using vortex mixer for 2 min and the emulsifying indices were measured after 24 h using Eq. 1. |
| Statistical data analysis |
| For statistical analyses, all the experiments were run in triplicates. The averages, standard deviations and analysis of variance (ANOVA), at 95% confidence level, were computed by using Microsoft Excel 2010 version. The standard errors for mean values, at 95% confidence level, were calculated and represented as error bars in all the graphical representations. The least significant difference (LSD) among the hydrocarbons tested was calculated by using Statistix software (Version, 8.1). |
| Results and Discussion |
| The most common property of all the biosurfactants is that they reduce surface and intra-molecular forces of heterogeneous systems by partitioning at air-liquid, liquid-liquid or solid-liquid interface. Such a property of biosurfactants helps them to reduce the surface tension and convert highly immiscible hydrocarbons into small droplets or micelles, and increase their bioavailability. These micelles are then easily taken up and utilized for growth and metabolism by microorganisms. Therefore, it is experienced that hydrocarbon contaminated sites are ideal sources for the isolation of biosurfactant producing microorganisms. Many scientists have recently reported variety of microorganisms having capability to produce biosurfactants from oil contaminated environments [2,32-37]. The bacterial strains isolated from oil contaminated sites are more prone to biosurfactant production. |
| Consequently in current study, the bacterial isolate P. putida SOL-10, from oil contaminated soil, was screened and optimized for biosurfactant producing ability. The physiological and biochemical characteristics of SOL-10 are given in Table 2. Based on negative starch, lipids and casein hydrolysis tests and positive catalase, oxidase, indole and citrate utilization, and lactose fermentation tests, the strain was tentatively identified as Pseudomonas putida. The API results obtained later on by using API software also confirmed that the isolate has 99% homology with P. putida strains. To further confirm the identity of SOL-10 strain, 16S rRNA sequencing was performed. The nucleotide sequence obtained after necessary trimming was 901 bp in size. The similarity search by BLAST program revealed maximum similarity (~98%) was with P. putida strains (Figure 1). Therefore the strain was named as P. putida SOL-10 strain. The partial nucleotide sequence has been submitted into online NCBI GeneBank repository and is available under accession number (JX534510). |
| A positive screening test (5.5 ± 0.2 cm, oil repellence zone) indicated that the isolate SOL-10 could be a suitable candidate for biosurfactant production. Figure 2 shows the gel image of the amplified product of rhamnosyltransferase l (rhlB) gene (723 bp) and compared with 1kb molecular weight DNA marker and another rhamnolipid positive isolate of P. aeruginosa isolated during current study (unpublished data).Most of the research documented on the production of rhamnolipids has been focused mainly on rhamnolipids of P. aeruginosa strains [38,39], whereas, very little studies have been conducted on P. putida so far [40-42]. Different studies reported the presence of unidentified rhamnolipid congeners in P. putida [40,43]. Similarly, in current study, we identified arhlB gene positive P. Putida isolate from oil contaminated soil. |
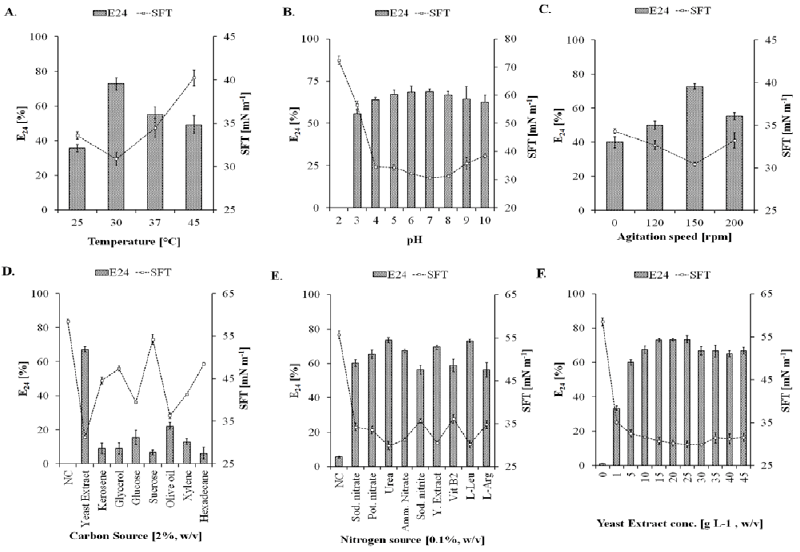
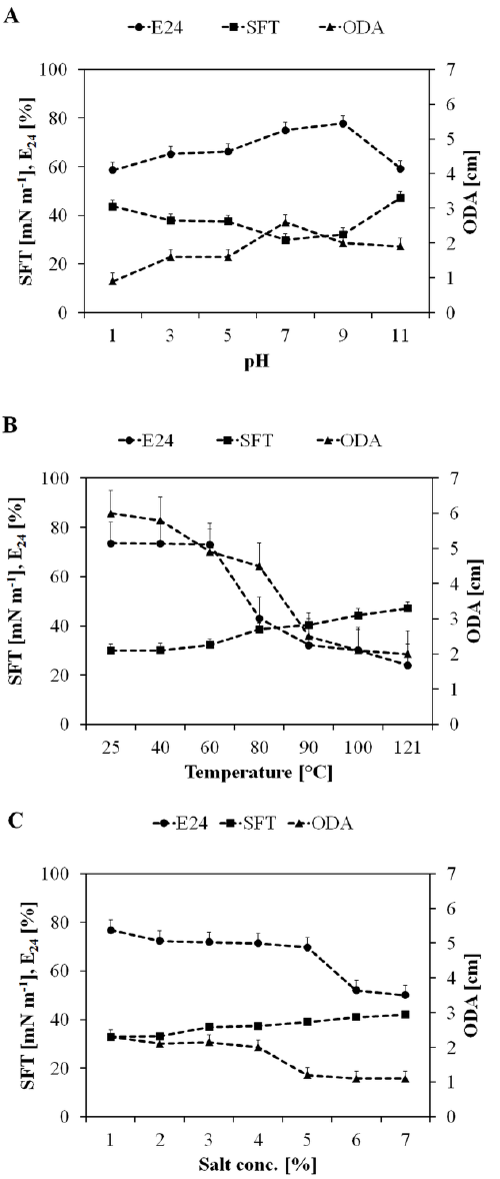
| On the basis of primary screening results, SOL-10 isolate was further investigated for the effect of culture conditions like temperature, pH, agitation, and carbon and nitrogen sources that usually effect growth and biosurfactants production [44]. Figure 3 shows the effect of different culture conditions on the biosurfactant-producing abilities of SOL-10, wherein the maximum biosurfactant production was achieved at 30°C (SFT, 30.89 ± 0.76 mN m-1; E24, 72.73 ± 3.5%) after 48 h of incubation. Although, the strain SOL-10 did not show significant biosurfactant production at 25, 37, and 45°C represented by 35.76 ± 2.1, 55.0 4.4 and 49.09 5.2%emulsifying activities, respectively (Figure 3A). However, significant reduction in surface tension of the cell-free culture broth was also achieved at 25, 37, and 45°C [SFT (mN m-1)=33.57 ± 0.44, 34.45 ± 1.1, 40.26 ± 0.93, respectively]. In general, the isolate revealed significant growth and biosurfactant production in the range of mesophilic temperature, i.e. 30 to 40°C, while a notable decrease in biosurfactant production was observed at higher and lower temperature ranges. This could be due to depressing effects of temperature on the biomass, E24, interfacial tension (IFT) and critical micelle dilutions [45]. The change in the pH of production medium also affected the biosurfactant production by P. putida SOL-10 strain. The strain did not show growth as well as biosurfactant production at pH 2.0, however, the maximum biosurfactant production was observed in the pH range 6-8 (E24, 69%; SFT, 30 to 32 mN m-1) after 72 h of incubation and only slightly declined (E24, 62.5%; SFT, 38.44 ± 0.9 mN m-1) during 120 h, while considerable activities were also detected in samples with pH range of 3.0 to 10.0 (Figure 3B). The pH tolerance at wide range may be attributed to the ability of the bacterium to produce alkaline byproducts, which in turn shifted the pH of the medium towards alkaline side (Figure 4). This is one of the reasons for growth and biosurfactant production by P. putida SOL-10 strain when grown at lower pH limits. The characteristic might in future be helpful in application of the strain and the product in leather, paper, textile and other industries working on variable pH ranges. |
| The cultivation broth showed maximum biosurfactant production at 150 rpm (E24, 72.73 1.5%; SFT, 30.42 0.2 mN m-1), while it remained comparatively lower at 120 rpm and 200 rpm (E24, 50 2.4%; SFT, 32.66 0.5 mN m-1and E24, 55.23 2.2%; SFT, 33.2 0.9 mN m-1, respectively) during 48 h. Similarly, fermentation experiments for biosurfactant production by P. putida strains at vigorous shaking speed have also been reported [40,46]. In the current study, the bacterial strain was also found capable of producing biosurfactants under static condition (0 rpm), giving an E24 of 40% after 48 h (Figure 3C). The absence of agitation resulted in extended lag phase, short exponential and long stationary phases (data not shown), which may be due the insufficient oxygen level than normally required during the initial acclimatization of the bacterium in growth medium. The higher agitation rate also negatively influenced the biosurfactant production possibly due to shear stress that would have resulted in detrimental effects on biosurfactant yield and growth kinetics of the bacterium. Our findings of optimum temperature, pH and agitation are somehow concurrent with previous works on the production of biosurfactants by different strains of P.putida that showed maximum production on 28C, pH 7.0 and 150 rpm [40,41,46]. The best results for rhamnolipids production from Pseudomonas species have been reported with the addition of either oils or residues rich in oils to the culture medium [29,47-49]. |
| Subsequently, the effect of different carbon and nitrogen sources was investigated (Figure3D-3F). Maximum biosurfactant production was observed in the medium containing 1.5-2.5% (w/v) yeast extract as carbon source (E24, ~72%; SFT, 29.95 mN m-1), however, varying concentration of yeast extract was found to have little effect on the biosurfactant production (Figure 3D and 3F). The strain demonstrated biosurfactant production with all the nitrogen sources tested. Figure 3E is showing the effect of organic and inorganic nitrogen sources on biosurfactant producing ability of P. putida SOL-10 isolate. The best emulsifying activities (E24, 73%) were detected when urea and L-leucin were used as nitrogen sources at a conc. of 0.1% (w/v) along with yeast extract (conc. 1.5%, w/v), as compared to inorganic nitrogen sources such as sodium nitrate, ammonium nitrate and potassium nitrate at 30C, 150 rpm and pH 7.0, showing maximum biosurfactant production. Likewise, none of the tested nitrogen sources displayed detrimental effects on growth as well as the biosurfactant production, which was really interesting. Although, the use of yeast extract resulted in significant biosurfactant production (E24, 72.33 1.3%; SFT, 30.54 0.25 mN m-1), it was found that supplementation of urea or L-leucin into the production medium maximized the productivity (E24, 73.45 ± 1.6% and 73.15 ± 1.0%; SFT, 29.91 ± 0.95 and 30.24 ± 0.87 mN m-1, respectively) as compared to nitrates of sodium, ammonium and potassium (Figure 3E). |
| The introduction of compounds containing amine groups like yeast extract and urea usually triggers the biosynthesis of either directly the peptide-containing biosurfactants or the enzymes regulating the biosynthesis of other biosurfactant types. Yeast extract is obviously the whole cell extract of yeast cells. It majorly contains proteins, short chain peptides, carbohydrates, free amino acids and nucleic acids. Therefore, it is supposed to be a rich source of not only carbon and nitrogen but also some essential trace elements as well. It was also therefore concluded that yeast extract alone can be utilized as a good source of nitrogen as well as carbon, which provided sufficient carbon and nitrogen content required by the isolate for biosynthesis of metabolic products. The production of biosurfactant by using amino acids L-Leucin and L-Arginine as nitrogen sources may be due to the direct uptake of amino acids as precursors for the surfactant biosynthesis, improving the biosurfactant yield [50,51]. The ammonium salts and amino acids might have further been incorporated into the biosurfactant’s primary structure resulting in positively charged functional groups making them cationic in nature, which shifted pH of the medium to alkaline side [52]. This might have enabled the P. putida SOL-10 to tolerate a wide range of pH for growth and metabolite production. Similar results were obtained with P. putida ML2 when nitrogen was supplied at the oxidation level of ammonia (urea) than with nitrogen provided as nitrate, although the strain was also able to assimilate nitrates [41]. |
| Previous work on biosurfactant production emphasized on the optimization of cultural conditions, because such factors could affect both the amount and type of biosurfactant produced [41,45,51]. In another study [53], biosurfactant production from gamma-ray induced P. putida 300-B mutant was carried out using distant carbon sources (viz. hydrocarbons, waste frying oils ‘WFOs’, vegetable oil refinery wastes and molasses). This was also one of the reasons to use olive oil, glycerol, kerosene, xylene and hexadecane as possible carbon sources for the production of biosurfactants from P. putida SOL-10 in current study. But in case of P. putida SOL-10, it was found that these hydrocarbons (except yeast extract) did not play a significant role in the production of biosurfactants. The only emulsification activity (E24, 25%) was found when olive oil was used. This may be due to their high hydrophobic lipid contents [51]. Although, we did not check the effects of different concentrations of those hydrocarbons, further studies are under the way for other factors that could affect growth as well as biosurfactant production. However, the maximum emulsification activity in samples with olive oil (E24, 25.37%) after 48 h have illustrated that it could be used as an additional medium supplement for the growth and as inducer for biosurfactant production by the strain. It was observed that olive oil and glucose could be used as inducers of growth and biosurfactants production by P. putida SOL-10 along with yeast extract (Figure 3D). Similar results were also reported with soybean WFO (waste frying oil) as carbon source and glucose as growth initiator under fed-batch cultivation [49]. The production of biosurfactants by P. fluorescens strain using substrates such as n-hexadecane, olive oil and glucose with an emulsifying activity of 49% when olive oil was used as the carbon source has also been reported [29]. |
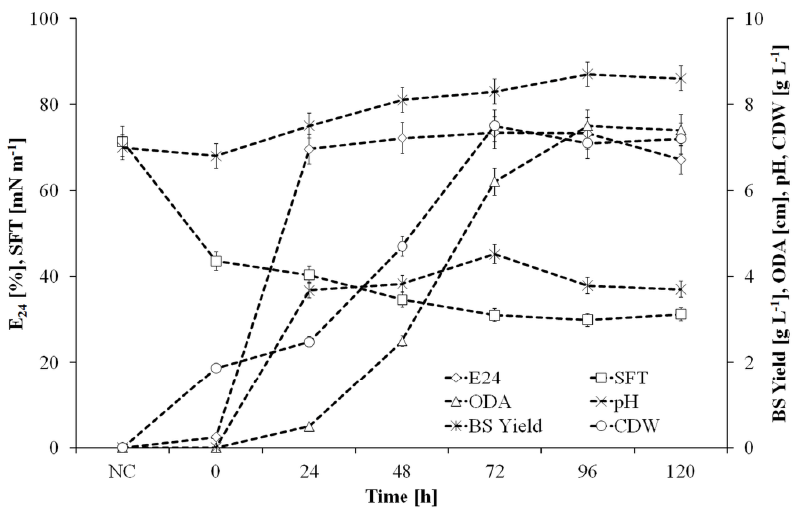
| Figure 4 shows the growth and biosurfactant production kinetics of SOL-10 isolate under optimized conditions in 2L shaking flak fermentation setup, where a growth associated production of biosurfactants was observed that declined during late stationary and death phase. It might be due to the enzymatic hydrolysis and uptake of these molecules because of substrate scarcity in the medium, or the inhibitory effect of biosurfactants on cell growth above a certain concentration, which induce subsequent degradation of these molecules [54]. The partial characterization of the biosurfactant produced under optimized conditions was performed by measuring the surface tension, pH, oil displacement zone and emulsification index (E24, %) in relation to biosurfactant yield and cell-dry weight. The surface tension was determined against a standard of de-ionized water (72 mN m-1). The surface tension of the cultivation broth started decreasing from the very first day (40.03 mN m-1) and reduced to 29.9 mN m-1 during 96 h. Similarly, the emulsifying activity of CFS reached to 73% after 72 h of incubation, which also confirmed the biosurfactant production by the strain. Thereafter, the surface tension slightly increased and emulsification index of the CFS declined that illustrated the depletion of biosurfactant from the cultivation broth after 120 h. The decline in biosurfactant level might be due to the inhibitory effect of biosurfactant on cell growth above certain concentration, which induced subsequent degradation of the molecules [54]. Another explanation could be the enzymatic hydrolysis and uptake of these molecules by the cells due to substrate scarcity in late growth phases of bacteria. The pH of the medium shifted to alkaline side with increasing concentration of the biosurfactant, decreasing surface tension and increasing emulsifying activity that indicated the cationic nature of the biosurfactant (Figure 4). The ability of biosurfactant to dissolve in water and alkalinity also indicated the presence of hydrophilic portion (i.e. carbohydrate, amino acid, phosphate or cyclic peptide) in it [55]. |
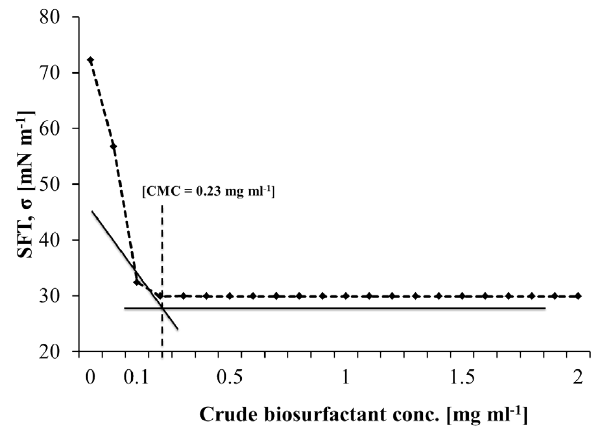
| The cell-free broth was then analyzed for critical micelle dilution (CMD) in order to quantify the biosurfactant concentration and to identify the optimum harvesting time for product recovery. Table 3 shows the time dependent relationship of surface tension and CMD of the cell-free broth at 3-fold dilutions (i.e. 10x, 100x, 1000x). It was observed that the surface tension of the CFS abruptly changed with dilution during initial 48 h of the incubation time, however, it was stable later on. This indicated enrichment of the production medium with significant amount of biosurfactant after 48 h of the incubation. The lowest surface tension representing most significant biosurfactant concentration was thus achieved during 72 to 96 h of incubation (Table 3). The critical micelle concentration (CMC) of the crude biosurfactant after recovery from fermentation broth was found to be in the range of 230 mg L-1 (w/v). The rhamnolipids from different bacteria have been reported to have their CMC values in the range of 10-600 mg L-1 [56,57]. |
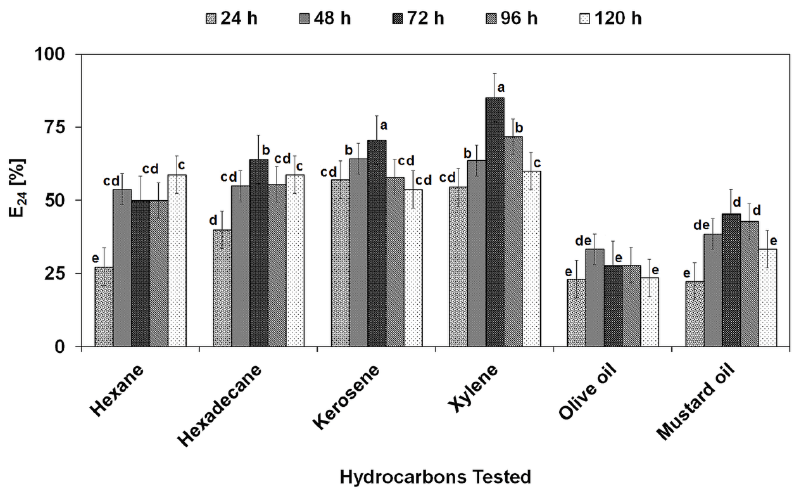
| After recovery from fermentation broth, the biosurfactant was investigated for its stability over a broad range of pH, temperature and salinity (Figure 6A-6C). Figure 5A shows that the biosurfactant was stable at broad pH range having maximum activity at pH 7 (E24, 75%, SFT, 34.9 mN m-1, ODA, 2.6 cm). At the lower pH range maximum activity was found at pH 5, while at the alkaline pH range activity was high at pH 9. In general, there was no significant loss of activity at a pH range of 1-11. The biosurfactant produced by P. putida SOL-10 was also found to be thermostable, where the surface and emulsifying activities were maintained even at the higher temperature of 100C and 121C, while the maximum activity (E24, 73%, SFT, 33.1 mN m-1, ODA, 1.7 cm) was observed at the moderate temperature range, i.e. 40°C (Figure 6B). Little changes were observed in the activity of biosurfactant between1-7% salt concentrations (Figure 6C). The results of this study were concurrent with previous reports about the stability of biosurfactants produced by P. fluorecens, P. aeruginosa MA01 and B. cereus NK1 [2,29,58]. The results were also found in accordance with the biosurfactant isolated from AlcanivoraxdieseloleiB-5 [59]. The stability of biosurfactant in the present work over a wide range of environmental conditions suggests that it maintains its activity at extreme conditions. A variety of hydrophobic sources i.e., n-hexane, n-hexadecane, kerosene, xylene, olive oil and mustard oil were evaluated in order to determine the emulsifying capability of the biosurfactant produced by P. putida SOL-10. Table 4 shows the statistical analysis of variance (ANNOVA) of emulsifying ability for all the hydrophobic sources tested. The results revealed that the biosurfactant has significantly higher (p<0.05) emulsifying capability against xylene and kerosene (>85% and ~70% respectively), as compared to other hydrophobic sources used (Figure 7). |
| The P. putida SOL-10 is a potential soil-borne, non-pathogenic biosurfactant producing strain. The optimization results concluded the selective behavior of the strain for biosurfactant production under favorable conditions. The preference of selective carbon and nitrogen sources at certain concentrations, as well as at specific environmental conditions demonstrated the importance of optimization studies in such cases. Although, yeast extract is mostly considered as nitrogen source, it also has sufficient amount of carbon that served as sole carbon and energy source for SOL-10 isolate during current study. However, due to expensiveness of yeast extract, further investigations are needed to replace yeast extract with alternate cheaper substrates for the production of surface active metabolites by the isolate, in order to make the process cost-effective as well. The current study would certainly aid some important knowledge to the existing data about biosurfactant production by different microorganisms, and the hydrocarbon emulsifying property of the biosurfactant can be helpful for future applications like bioremediation of contaminated environments. |
| Acknowledgements |
| The authors are highly grateful to Higher Education Commission (HEC), Government of Pakistan, for funding this study under HEC-Project No. 1366, entitled “Production and characterization of biosurfactants produced by indigenous microorganisms. The authors have no conflict of interest to declare. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15137
- [From(publication date):
August-2013 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 10408
- PDF downloads : 4729
