Review Article Open Access
Wildlife Zoonoses
David T. S. Hayman*
Cambridge Infectious Diseases Consortium, Department of Veterinary Medicine, University of Cambridge, Madingley Road, Cambridge, CB3 0ES, UK
Department of Biology, Colorado State University, Fort Collins, Colorado, CO80523, USA
Animal Health and Veterinary Laboratories Agency, Wildlife Zoonoses and Vector-borne Diseases Research Group, Department of Virology, Veterinary Laboratories Agency – Weybridge, Woodham Lane, Weybridge, New Haw, Addlestone, Surrey, KT15 3NB, UK
- *Corresponding Author:
- David T. S. Hayman
Department of Biology
Colorado State University, Fort Collins
Colorado, CO80523, USA
E-mail: dtsh2@cam.ac.uk
Received date: October 05, 2011; Accepted date: December 08, 2011; Published date: December 20, 2011
Citation: Hayman DTS (2011) Wildlife Zoonoses. Epidemiol S2:001. doi:10.4172/2161-1165.S2-001
Copyright: © 2011 Hayman DTS. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Epidemiology: Open Access
Abstract
Infectious diseases are still to be found among the top causes of human deaths globally. The majority of human pathogens are zoonotic and many have their origins in wildlife. The cost of new infections to societies in terms of human mortality and morbidity can be enormous. Humans have contact with vastly more infectious agents of wildlife origin than spillover and emerge in human populations. Therefore, understanding and predicting zoonotic infection emergence is complex. Changes in the ecology of the host(s), the infection or both, are thought to drive the infection emergence in a range of different host-infection systems. Here key recent studies regarding how changes in host ecology, receptor use and infection adaptation relate to spillover and emergence from wildlife reservoirs are reviewed. The challenges wildlife zoonoses pose to epidemiologists are also discussed, along with how developments in technology, such as PCR, have changed perspectives relating to wildlife as hosts of zoonotic infections.
Introduction
In the late 1960s and 1970s, many people working in public health in industrialized societies such as the USA believed that infectious diseases would cease to be a major health threat [1]. Vaccines existed for some of the most devastating diseases, including poliomyelitis, measles, and smallpox, and malaria had been eradicated from large regions, including Europe [2,3]. In 1977 the last case of smallpox was reported and it became the first infectious disease to be eradicated globally. Rabies vaccines, which had existed since Pasteur in 1885, had successfully been trialled in an oral bait delivery system, and signs were that one of the most feared animal infections could be controlled [4].
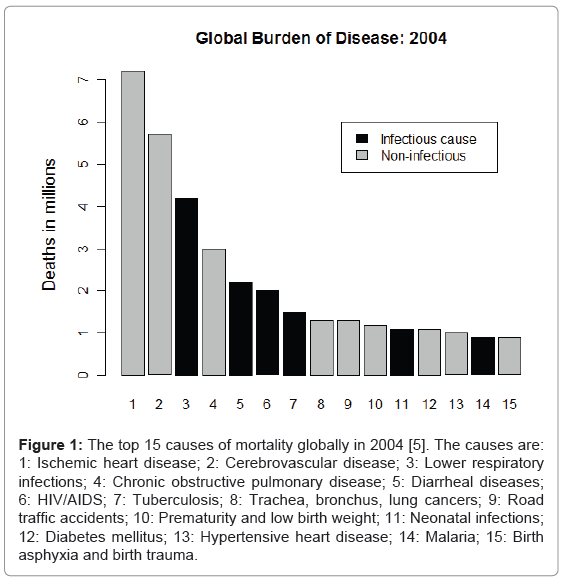
However, it is now well into the 21st Century and infectious diseases are still to be found among the top causes of human deaths globally [5] (Figure 1). The majority of human pathogens are now recognized to be zoonotic [6-8]. Zoonotic infections are those of non-human origin that infect humans, and most zoonotic infections are of wildlife origin [6-8].
Figure 1: The top 15 causes of mortality globally in 2004 [5]. The causes are: 1: Ischemic heart disease; 2: Cerebrovascular disease; 3: Lower respiratory infections; 4: Chronic obstructive pulmonary disease; 5: Diarrheal diseases; 6: HIV/AIDS; 7: Tuberculosis; 8: Trachea, bronchus, lung cancers; 9: Road traffic accidents; 10: Prematurity and low birth weight; 11: Neonatal infections; 12: Diabetes mellitus; 13: Hypertensive heart disease; 14: Malaria; 15: Birth asphyxia and birth trauma.
Here I review the history and impacts of emerging infectious diseases in today’s society, and processes of infection emergence from wildlife. The importance of factors relating to host ecology, receptor usage and host range, and pathogen adaptation in spillover and emergence into human populations will be discussed. Finally, recent advances in technology and the challenges emerging zoonoses pose for epidemiologists will be highlighted.
History of Emerging Infectious Diseases
It is now generally recognized that even the most “human” of infectious diseases at some time had their origins in other animals, typically wild, but also domesticated [8]. For example, well-known Old World human pathogens that now only infect humans, such as measles and smallpox, have animal origins [8,9]. Measles is thought to have derived from spillover of rinderpest or canine distemper virus infection in Mesopotamia following urbanization circa 8000BCE and human population sizes reached a threshold size that allowed persistence [10-12]. The increase in human population density and today’s increase in connectivity and encroachment into wild areas probably means that we are more susceptible to infection emergence, and subsequent disease, than ever [13,14].
Numbers and Costs of Emerging Zoonoses
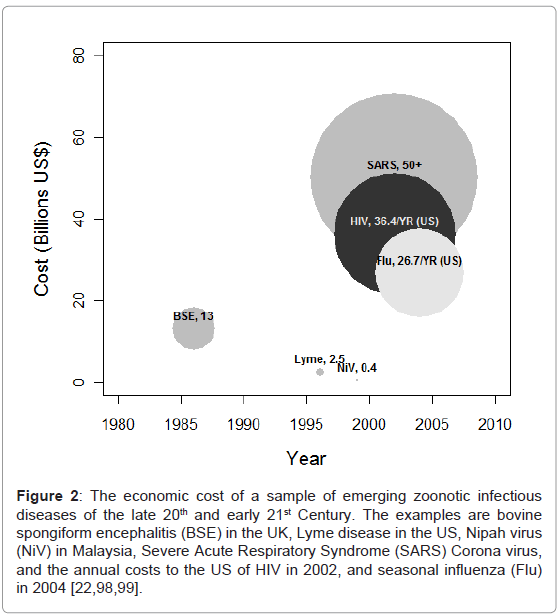
Relatively recent reviews have estimated that approximately 175 of the 1400 human pathogen species recognized are emerging or reemerging, and between 58-75% of all infections are zoonotic [7,15]. The cost to humans of these emerging infections can be substantial, both in terms of lives and economics (Figure 2). A pertinent example is the emergence of human immunodeficiency virus (HIV), which occurred due to bush meat hunting of simian immunodeficiency virus (SIV) infected primates in Africa in the early part of the 20th Century [16-18]. Indeed, many groups of HIV-2, a relative of an SIV that infects sooty mangabeys (Cercocebus atys atys), still largely circulate only in African human populations, and have yet to become pandemic infections [19,20]. In 2009 the WHO estimated 33.3 million people lived with HIV infection, 2.6 million new HIV infections occurred and 1.8 million people died due to acquired immune deficiency syndrome (AIDS) related illnesses [21]. New HIV infections in the United States in 2002 alone were estimated to cost $36.4 billion per annum, including $6.7 billion in direct medical costs and $29.7 billion in productivity losses [22].
Figure 2: The economic cost of a sample of emerging zoonotic infectious diseases of the late 20th and early 21st Century. The examples are bovine spongiform encephalitis (BSE) in the UK, Lyme disease in the US, Nipah virus (NiV) in Malaysia, Severe Acute Respiratory Syndrome (SARS) Corona virus, and the annual costs to the US of HIV in 2002, and seasonal influenza (Flu) in 2004 [22,98,99].
Infection emergence is complex because many species share infections and jumps between host species are common and natural. Recent estimates suggest that there may be 8.7 million eukaryote species on earth [23]. It is likely that each of these has a suite of viral infections, many of which they may share with other species. Given the number of wildlife species humans have contact with around the globe; the exposure rate of humans to infectious agents vastly exceeds the rate of spillover from wildlife to humans. The many orders of magnitude difference between exposure and spillover rates make predicting emergence difficult. There have, however, been several efforts aimed at predicting infection emergence, both relating to the likely infections or regions of the globe where new emergence events may occur. Jones et al analyzed data to predict likely hot-spots for infections to emerge from, whereas other efforts have aimed to determine from which animal species or orders infections emerge [7,13,15,24]. Further analyses have identified that viral traits can predict emergence [25,26].
Emergence of an infection may be defined as the recent increase in the geographical and/or host range or the incidence of the infection [27-29]. Increasing human populations and encroachment into wildlife habitats are thought to particularly increase the chance of emergence of infections from wildlife hosts, whilst global climate change may now be playing a role in changing pathogen range [30-32]. Changes in the ecology of the host(s), the infectious agent, or both, are thought to drive the infection emergence in a range of different host-infection systems [26,27,33,34]. However, frequently infections that are classified as emerging are often single spillover events with individual human infections [35].
Recent reviews have provided useful classifications for infections at various stages of the process of infection emergence from wildlife to humans [6,8]. The process is difficult to predict when considering specific infections, but generally formulaic, and can be described by several stages (1 through 5) of transmission between the infectious agent’s original reservoir(s) to a new host. Reservoirs can be thought of in relation to the target population. Haydon and others defined reservoirs as “one or more epidemiologically connected populations or environments in which the pathogen can be permanently maintained and from which infection is transmitted to the defined target population”, the target being humans in the case of zoonotic infections [36].
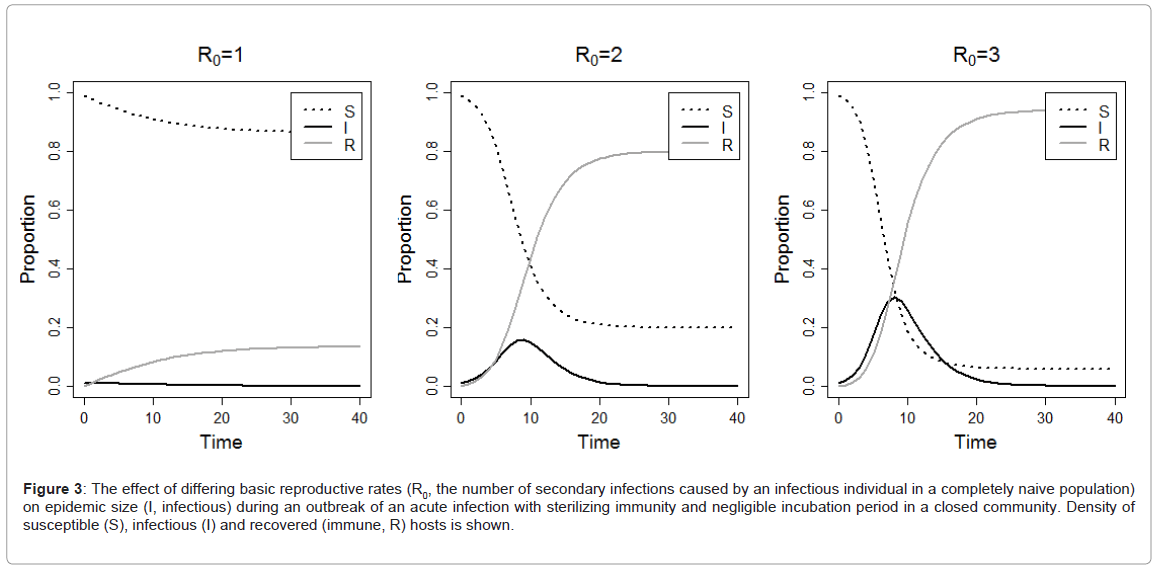
By definition, endemic transmission occurs within reservoir populations, so called stage 1 infections [6,8]. If there is frequent exposure between species or locales (sometimes referred to as ‘chatter’), some individual spillover infections may occur, and these are called stage 2 infections. Stage 2 infections include those such as rabies virus (RABV) and West Nile virus (WNV) [6,8,37,38]. The epidemiological parameter R0 represents the number of secondary cases acquired from a primary infection in a completely susceptible population. For an infection to invade a population, R0 must be greater than one (Figure 3), whereas if R0 is below one the infection will rapidly disappear from the population. If infection replication in the new host leads to transmission, and if R0 is less than one, there may be limited local spread (sometimes called stuttering chains), but no persistence [6]. These are called stage 3 infections, and examples include infections such as Nipah virus (NiV) and monkeypox [39-41]. Once the transmission chain is long enough and R0 greater than one, establishment of infection in the new species or locale can occur [6,42,43]. These stage 4 infections are able to cause prolonged outbreaks of infection in the human populations and examples include Yellow fever virus (YFV) and Dengue virus (DENV). Obviously, contact rates and host population sizes need to be great enough for infection to become endemic in the new host population (stage 5 infection, such as HIV) and adaptation may occur [6,11,26].
Figure 3: The effect of differing basic reproductive rates (R0, the number of secondary infections caused by an infectious individual in a completely naive population) on epidemic size (I, infectious) during an outbreak of an acute infection with sterilizing immunity and negligible incubation period in a closed community. Density of susceptible (S), infectious (I) and recovered (immune, R) hosts is shown.
Wildlife as Reservoirs of Emerging Infections
As discussed above, wildlife is the reservoir of a number of zoonotic infections. In the Cameroon-Congo Basin, for example, infections originating from wildlife have included HIV-1 and 2, monkeypox, arboviruses (YFV and DENV), anthrax, Ebolavirus (EBOV), and human T-lymphotrophic viruses (HTLV 1 and 2) [18,31,41,44-46]. Bushmeat hunting is thought to have led to the emergence of both HIVs and HTLVs in the human population in the region. Bats have been linked to a number of fatal zoonotic RNA infections, including henipaviruses (Hendra-HeV and NiV) in Asia and Australia [47,48], filoviruses (Marburgvirus; MARV, and EBOV) in Africa and Asia [45,49], SARS-like coronaviruses (CoV) [50] in Asia and lyssaviruses (including RABV) globally [51]. The majority of viruses isolated from bats have been RNA viruses. RNA viruses typically have high replication and mutation rates due to their low proof reading ability, and large population sizes. These factors potentially allow rapid adaptation in new hosts, which is the hypothesized mechanism to explain why so many recognised emerging infections are RNA viruses [28,29,42].
Ecological or adaptive bridging may facilitate zoonotic emergence from wildlife. Bridging hosts may be a domestic or wild species. Examples of both ecological and adaptive bridging can be found when examining emergence events of zoonotic viruses with bat origins. Primates, for example, are thought to be bridging hosts between the fruit bat reservoir and humans in EBOV outbreaks in Central Africa [52]. Himalayan palm civets (Paguma larvata) possibly acted as adaptive bridging hosts between the putative reservoir, Rhinolophid (Horseshoe) bats, and human populations in the SARS-coronavirus pandemic in 2002-03 [53]. Henipaviruses (paramyxoviruses), which can cause fatal encephalitis in man, have Pteropus fruit bats as their putative reservoir hosts and domestic animals as bridging hosts; the transmission of HeV to humans has occurred through horses and the transmission of NiV has occurred through pigs [54-56]. In Malaysia in 1999, over one million pigs were culled to control a NiV outbreak, which killed 105 people. More worryingly for NiV, direct bat-tohuman transmission, followed by human-to-human transmission of NiV is thought to have occurred in Bangladesh [39,57,58].
Host Ecology and the Causes of Virus Spillover into Humans
The transmission dynamics of HeV in Australian Pteropid bats were studied by Plowright et al using a mathematical model parameterised with field and laboratory data [59]. They found evidence of decreasing migration of bats in Australia, possibly due to urbanisation and changes in food availability, which may have influenced the strength of HeV epidemics. Decreased migration appears to be creating more intense HeV outbreaks after local viral reintroduction due to decreased migration leading to reduced transmission between colonies and thus colony immunity. This loss of immunity is hypothesized to lead to increased HeV epidemic size once infection was re-introduced into the colony. Hence the reduced seasonal migration of the bats is believed to increase in the strength of the seasonality in viral dynamics and the size of the epidemics in the bat colonies, thus increasing the chance of spillover.
Duke-Sylvester et al have modelled the effects of seasonality in raccoon rabies and found that strong seasonality in birth pulses produces spatial asynchrony in the outbreak of infectious diseases [60]. These findings are important for surveillance and control programmes, allowing better targeted vaccination, as well as increasing our understanding of the drivers of infection dynamics in their wildlife hosts. Indeed, rabies virus is an exemplar of a stage 2 infection that causes individual human cases and no human to human transmission (R0=0), but one which provides numerous well studied examples that highlight the difficulties in controlling emerging infections from wildlife.
Rabies remains the only disease considered to have a 100% mortality rate and dog rabies is estimated to cause over 55,000 human deaths globally per annum [61]. Despite originating in bats, RABV is now enzootic throughout much of the world with the domestic dog (Canis familiaris) being the principal vector [61]. However, RABV is distributed worldwide in mammalian carnivores, with RABV variants undergoing genetic evolution in particular host(s), leading to new clades or biotypes. Rabies in wildlife has led to three dramatic post- World War II epidemics in the 20th Century. These epidemics were in foxes (Vulpes spp.) in both Europe and North America, starting in the 1940’s, and raccoons (Procyon spp.) in North America, starting in the mid-1970’s [62,63]. Wild terrestrial carnivore reservoirs vary according to those carnivores present in the local fauna, but include skunks (Mephitidae), foxes, and raccoons in North America [64]. The virus-host reservoir relationships mean that despite control of domestic dog rabies, terrestrial rabies can persist or increase. These findings highlight the problems that exist when there are multiple hosts and when the ecology of the reservoir or reservoirs makes control difficult. Control of RABV in dogs has prevented many human rabies cases in developed countries, and more recently that control has been extended to terrestrial wildlife. Unfortunately, control of terrestrial rabies is still lacking in large parts of Africa and Asia. The number of domestic and wild animals which have succumbed to rabies is impossible to estimate, however, Knobel et al. [61] estimated 5,200,000 dogs are killed each year in Africa and Asia alone, simply as part of rabies control programmes. However, the presence of numerous bat reservoirs mean that complete control is unlikely and although few in number, bat-derived rabies now cause the greatest number of human deaths from rabies in the USA. In 1993, a small outbreak of rabies cases occurred in foxes on Prince Edward Island, Canada. The genetic detection of a bat RABV variant within this fox population was confirmed and through detection of low levels of virus in salivary gland material it was assumed that some degree of intraspecific transmission among the foxes had occurred [65]. A second report from Arizona, USA in 2001, highlighted the infection of a number of skunks with a bat RABV variant [66].
The numerous emerging zoonoses of wildlife origin which have vector-borne life cycles pose another substantial challenge. Well known examples of wildlife derived arthropod borne infections include malaria causing Plasmodium, DENV, YFV and Borrelia burgdorferi. Borrelia burgdorferi, the bacterium that causes Lyme disease, is spread through the bite of infected ticks. Blacklegged ticks (or deer tick, Ixodes scapularis) spread the disease in the northeastern, mid-Atlantic, and north-central United States, and the western blacklegged tick (Ixodes pacificus) spreads the disease on the Pacific Coast, however, the incidence of infection has been steadily increasing [67]. Over 20,000 Lyme disease cases were confirmed in the USA each year from 2002 to 2010, with the majority of cases in the northeastern USA [67]. The cause of this increase is unknown, however, it is possible that changing ecological processes, including tick hosts such as deer, and increased human-tick exposure are to blame [68].
Changes in host ecology can lead to profound effects in disease and infection transmission. Now largely considered human infections, DENV and YFV had their origins in wild primates. The wildlife vectors of DENV and YFV, Aedes aegypti and Aedes albopictus, are mosquito species that have been spread globally from their native origins by human trade throughout the tropics [69]. Aedes aegypti originated from Africa and A. albopictus from South-East Asia [70]. Today, Dengue fever is the most rapidly spreading mosquito-borne viral disease in the world today. The incidence has increased 30-fold in the last 50 years, with an estimated 50 million dengue infections occurring annually [71]. Interestingly, it remains a largely sylvatic infection in West Africa [69,72].
Yellow fever virus, believed to have originated in Africa due to the genetic diversity detected among African viruses, is now endemic in South America with both sylvatic and urban life-cycles [70,73]. Endemic sylvatic YFV in South America means that eradication in now highly improbable.
Receptor Usage and Host Range
Infections evolve to adapt to their host and host specific receptors [74,75]. Some phenotypes, however, will be conserved and small mutations may lead to a broader receptor range than is normal. Some viruses use conserved receptors that are likely to have a broad host range. For example, henipaviruses are believed to use Ephrin-B2 and Ephrin-B3 receptors [76,77]. These are highly conserved receptors that are present in all animal species examined, potentially explaining the broad species range infected by these viruses. However, despite recent studies suggesting widespread evidence of henipavirus infection in potential reservoirs (such as African bats, Figure 4), spillover is currently only reported repeatedly in Bangladesh (to humans) and Australia (to horses) [39,57,59,78]. Questions remain, therefore, as to whether these findings are due to surveillance, ecological factors or factors relating to the specific viruses circulating in those regions. Findings such as henipaviruses utilizing such conserved receptors should remind us that while humans like to categorise themselves as being different from other animals, there exists approximately 80% homology between human and mouse genes with single identifiable orthologues [79]. Therefore, while there are many differences and major histocompatibility complex (MHC) receptors show huge plasticity enabling us to recognise an enormous suite of infections, it is clear many cellular features are conserved across Orders, and that parasitic infections themselves have evolved methods that enable them to circumvent the host immune responses.
Figure 4: Eidolon helvum roosting in Accra, Ghana. This widespread African bat species often roosts in urban areas in Africa, is a source of bush meat, and has recently been found to be infected with henipaviruses. However, human henipavirus infections remain unreported in Africa, whereas in parts of Bangladesh and Australia frequent spillover occurs. It is unknown whether this difference is due to surveillance, ecological or virological differences.
Many tick or arthropod-borne infections, such as the flaviviruses WNV, YFV, DENV and Tick-borne encephalitis virus, are already able to infect multiple hosts, including humans [80]. It appears that many of these are increasing their incidence in humans as we increase the human-vector contact. However, DENV and YFV appear to have adapted to humans so that viral replication in human cell cytoplasm is high enough and human-mosquito-human transmission occurs successfully. Interestingly, Pulliam and Dushoff analyzed viruses of domestic artiodactyls (e.g. pigs, sheep, cattle, and goats) and found replication without nuclear entry to be the strongest predictor of crossspecies transmission [25]. Therefore, understanding receptor use and site of replication may be useful predictors of disease emergence due to cross-species transmission. Furthermore, although the mechanisms are different for bacteria, it is unknown why only 12 of the 36 known Borrelia species cause disease in humans, but the use of different surface proteins may be a mechanism to explain the different pathogenesis of different Borrelia species. For example, cell surface proteins have been shown to be important determinants for cellular attachment and establishment of infection in human cells by B. Burgdorferi, the aetiological agent of Lyme disease [81].
Pathogen Adaptation
Most of the infections discussed above are generally defined as being at stages 2 through 4 in the emergence process. These are endemic infections in wildlife, but cause individual spillover infections in humans (stage 2, R0=0), through to those that cause chains of transmission in human populations (R0≤1, stage 3; R0>1, stage 4; Figure 3) [6,8]. Once there has been sustained human to human or humanarthropod vector-human transmission, there may be adaptation to the human before an infection becomes a human only infection (stage 5) [6,8]. A recent review has challenged how much evidence there is for adaptation being a prerequisite for emergence and transmission in new hosts [82]. The authors proposed four mechanisms for emergence; two with ecological drivers and no adaptation, and two with adaptive drivers when selection is required. The authors highlighted that little evidence exists for the importance of adaptation even for four well studied viruses that are known to have switched hosts (influenza virus, SARS-CoV, canine parvovirus and Venezualan equine encephalitis virus).
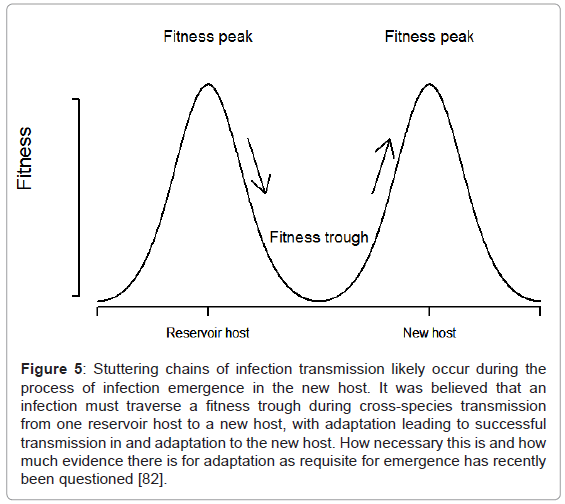
Classically, infections such as tuberculosis and malaria were thought to have co-evolved with humans (see below for a discussion), and RNA viruses, for example, were thought more likely to be emerging infections. The poor proof-reading ability of the RNA-dependent RNA polymerase and high virus populations meaning higher mutation rates are thought to be the mechanistic reasons for RNA viruses being more likely to be emerging infections [42,43,83]. These factors were thought to enable RNA viruses to move from one fitness peak (successful transmission in the reservoir host species) to another (successful transmission in the recipient host species) for emergence (Figure 5) [83]. If the traversing of such a fitness valley, during which time the infection is inadequately adapted to both species, is necessary, high rates of recombination or re-assortment in viruses, or lateral gene transfer in bacteria, are likely to further assist with this process. Again using rabies as an example, this highly promiscuous RNA virus is able to affect almost all mammals, suggesting adaptive bridging is less important for infection of a new host than ecological bridging for lyssaviruses, although RABV variants may adapt to their “new” hosts and a degree of host restriction exists [51,84]. Despite genetic analysis of glycoprotein sequences from lyssaviruses circulating in both bat and terrestrial carnivore species suggesting that host switching of lyssaviruses from bats to other mammals has occurred repeatedly and successfully in history, the majority of cases of spill-over events are likely dead-end infections [51]. Thus, the case for adaptation as a requisite for adaptation to a new host is still unclear [82].
Figure 5: Stuttering chains of infection transmission likely occur during the process of infection emergence in the new host. It was believed that an infection must traverse a fitness trough during cross-species transmission from one reservoir host to a new host, with adaptation leading to successful transmission in and adaptation to the new host. How necessary this is and how much evidence there is for adaptation as requisite for emergence has recently been questioned [82].
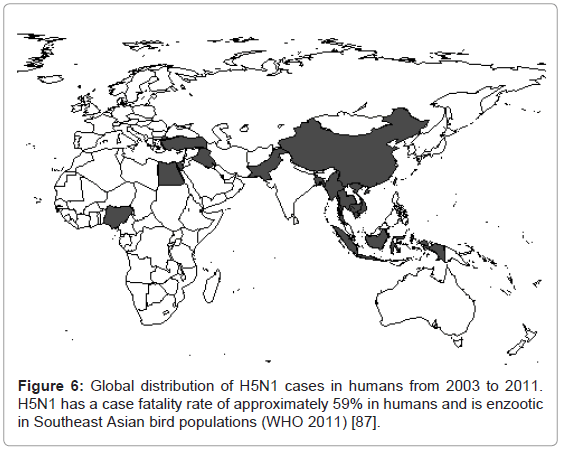
New influenza emergences, such as H1N1 in 2009, are classic examples of viral re-assortment [85]. Re-assortment speeds the process of change and putatively adaptation, thus potentially allowing crossspecies transmission, immune escape of the viruses and circulation in new populations of susceptible hosts. In the year after the emergence of H1N1 in 2009, the CDC estimated there were 43-89 million human cases, with 8870 to 18300 H1N1 related deaths [86]. Thus, the case fatality rate was actually low for human influenza A infection. Prior to the emergence of H1N1, concern was, and still is, regarding H5N1. The worry over this virus is that it has caused human fatalities in the Old World (Figure 6) and rather than the approximately 0.02-0.04% case fatality rate of the H1N1 pandemic of 2009, H5N1 infections between 2003 and 2011 have had a case fatality rate of approximately 59% [87]. It is unknown if adaptation is required for H5N1 emergence into the human population, although an absence of human to human transmission may suggest some adaptation to humans is necessary.
Examples of infectious agents readily able to recombine genetic material are found among the coronaviruses (the aetiological agent for SARS). The SARS-CoV outbreak is believed to have been due to an adaption event, possibly by recombination, of a bat-derived CoV, probably in an intermediate civet host, before transmission to humans [53]. However, as highlighted by Pepin and colleagues [82], it remains unclear whether adaptation processes were necessary for HIV, measles, and seasonal influenzas becoming endemic human-only infections following emergence and adaptation to humans from wildlife.
Recent Advances in Technology
The use of molecular tools has dramatically changed our understanding of the numbers of infections and roles different hosts play in emerging zoonoses. Scientists searched for several decades for the reservoirs of EBOV. Today it is widely recognized that bats play a key role in the infection dynamics of EBOV and MARV, another filovirus equally pathogenic to humans, due to the molecular detection of viral RNA from bat hosts, which preceded the more recent isolation of MARV from bats [45,49,88]. Interestingly, both filovirus sequences have been discovered in rodent and bat genomes as non-retroviral integrated RNA viruses, suggesting ancient viral circulation and integration [89].
After studies linked SARS-CoV to bats, efforts were made to sample different species of bats intensively. The enormous diversity of coronaviruses that have been found in bats in China alone and their relationship to CoVs infecting humans and a range of other species globally has dramatically altered the understanding of CoV diversity and ecology [90]. This increase in the known CoV diversity and the role of bats in CoV ecology is despite SARS-CoV never having been cultured in a laboratory. Equally as astonishing is the diversity of Astroviruses (AstV) detected by PCR in bats in Hong Kong [91,92]. The diversity of bat AstV and relationships with human and other animal AstV clearly suggest that bats play or have played an integral role in AstV ecology and evolution. But it is notable that these studies would have been limited if it were not for molecular tools.
Most recently, Plasmodium falciparum, the cause of several hundred million cases of clinical malaria and more than one million deaths annually, has recently been found in gorillas in Central Africa [93]. Prior to this finding, it was hypothesized that it diverged from a chimpanzee infection more than 5,000,000 years ago. This finding has transformed the current understanding of the origin of malaria in humans, suggesting that P. falciparum is of gorilla origin, not ancient human or chimpanzee. However, it required careful molecular and phylogenetic analyses to determine in which direction the transmission occurred due to the close phylogenetic relationships between human and gorilla P. falciparum.
The use of molecular tools, therefore, has been essential in increasing our understanding how humans and other animals share infections. The rapid current development of next-generation pyrosequencing (NGS) will likely increase this understanding further. NGS technology is increasingly driving the understanding of human genomes and the human gut microbiome. Studies using NGS approaches have begun to quantify the diversity of potentially infectious agents humans may be exposed. For example, NGS studies of human feces suggest humans consume, and therefore are exposed to, a range of genetically diverse plant viruses [94]. The likelihood of human disease and adaptation of these plant viruses to humans is unknown. However, NGS studies are likely to advance our understanding of the genetic diversity of infectious agents humans are exposed to and the processes of zoonotic spillover and emergence in the future.
Challenges for Epidemiologists
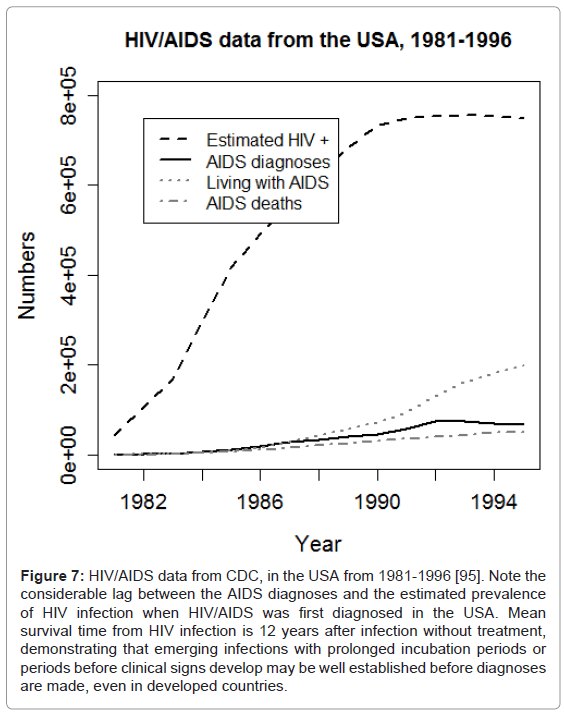
Despite the rapid improvement of diagnostic techniques and increase of laboratories and epidemiological training globally, substantial challenges still remain. Infections that emerge, such as HIV, that have long incubation periods or emerge in more remote or poor areas or countries may become established in human populations before surveillance systems are in place. For example, SARS-CoV was rapidly identified and controlled, but HIV infection was only diagnosed once AIDS was recognized in the USA, by which point the infection was well established in the human population globally (Figure 7) [95].
Figure 7: HIV/AIDS data from CDC, in the USA from 1981-1996 [95]. Note the considerable lag between the AIDS diagnoses and the estimated prevalence of HIV infection when HIV/AIDS was first diagnosed in the USA. Mean survival time from HIV infection is 12 years after infection without treatment, demonstrating that emerging infections with prolonged incubation periods or periods before clinical signs develop may be well established before diagnoses are made, even in developed countries.
Consequently, while epidemiological techniques will discover where the infections came from and who is at risk, the emergence process may be too late to prevent. A particularly interesting example is the increased incidence of monkeypox, a smallpox relative in the Orthopox genus of poxviruses, in Democratic Republic of Congo (DRC). From 1981–1986 to 2006–2007, monkeypox incidence increased from 0.72/10000 to 14.42/10000 respectively [40]. This increase is hypothesized to be due to a reduction in immunity due to the cessation of smallpox vaccination programme, and the continued exposure to wildlife reservoirs within the region. Epidemiologists discovered the increase in monkeypox incidence in human populations in DRC and further studies are required to understand the spillover and potential emergence of this infection in these human populations from wildlife. Further evidence of the problems relating to surveillance is demonstrated by a study by Mallewa and others in Mallawi [96]. In a malaria-endemic area of Africa, RABV was found to cause fatal central nervous system (CNS) infection in 14 (10.5%) of 133 CNS infection cases, with 3 (11.5%) of 26 fatal cases originally attributed to cerebral malaria due to RABV. In another study Mebatsion et al discovered 2 lyssaviruses of bat origin causing fatal dog infections in a sample of 113 rabies cases in Ethiopia [97]. Notably, the current vaccines do not provide protection against these particular bat-derived lyssaviruses, and differentiation of these non-RABV lyssaviruses requires molecular or monoclonal antibody techniques. These examples of problems relating to surveillance and diagnosis of well known infections demonstrate how understanding spillover and emergence processes are hampered by poor surveillance and lack of diagnostic capabilities. Thus, the examples here demonstrate the importance of ensuring good surveillance capabilities exist globally. Better surveillance systems are required throughout the world, because examples such as influenza and HIV remind us of how connected nations are and how devastating infection emergence can be.
Conclusion
In summary, to understand the infection dynamics that allow persistence in the reservoir species and the triggers that lead to emergence requires good knowledge of the reservoir species’ biology and ecology, as well as receptor usage and host range, and pathogen evolution probability. Understanding these will require multidisciplinary studies and pose unique challenges to epidemiologists. Epidemiologists will be required to work with ecologists, clinicians and diagnosticians throughout the world in order to understand better, and therefore predict and control, zoonotic emergence events from wildlife hosts.
Acknowledgements
Thanks to Mi Chen for help in obtaining the CDC HIV data, Angie Luis, Kim Pepin and an anonymous reviewer for comments on the manuscript, James Wood, Tony Fooks and Andrew Cunningham for discussions on wildlife zoonoses over the years, and Dr. Patrick Sullivan for the invitation to contribute to this issue.
David Hayman is funded by the Wellcome Trust (DTSH), the Research and Policy for Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security, Fogarty International Center, National Institutes of Health.
References
- Morens DM, Folkers GK, Fauci AS (2004) The challenge of emerging and re-emerging infectious diseases. Nature 430: 242-249.
- Feachem RG, Phillips AA, Hwang J, Cotter C, Wielgosz B, et al. (2010) Shrinking the malaria map: progress and prospects. Lancet 376: 1566-1578.
- Hinman AR (1999) Economic aspects of vaccines and immunizations. C R Acad Sci III 322: 989-994.
- Smith TG, Wu X, Franka R, Rupprecht CE (2011) Design of future rabies biologics and antiviral drugs. Advances in Virus Research 79: 345-363.
- http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.html
- Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JR, et al. (2009) Epidemic dynamics at the human-animal interface. Science 326: 1362-1367.
- Taylor LH, Latham SM, Woolhouse ME (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356: 983-989.
- Wolfe ND, Dunavan CP, Diamond J (2007) Origins of major human infectious diseases. Nature 447: 279-283.
- Pearce-Duvet JM (2006) The origin of human pathogens: evaluating the role of agriculture and domestic animals in the evolution of human disease. Biol Rev Camb Philos Soc 81: 369-382.
- Keeling MJ (1997) Modelling the persistence of measles. Trends in Microbiology 5: 513-518.
- Keeling MJ, Grenfell BT (1997) Disease extinction and community size: modeling the persistence of measles. Science 275: 65-67.
- Chastel C (1999) Viruses and civilization. Med Trop (Mars) 59: 425-429.
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, et al. (2008) Global trends in emerging infectious diseases. Nature 451: 990-993.
- Hosseini P, Sokolow SH, Vandegrift KJ, Kilpatrick AM, Daszak P (2011) Predictive power of air travel and socio-economic data for early pandemic spread. PLoS One 5: e12763.
- Cleaveland S, Laurenson MK, Taylor LH (2011) Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci 356: 991-999.
- Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, et al. (1998) An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391: 594-597.
- Zhu T, Ho DD (1995) Was HIV present in 1959? Nature 374: 503-504.
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397: 436-441.
- Gao F, Yue L, Robertson DL, Hill SC, Hui H, et al. (1994) Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. Journal of Virology 68: 7433-7447.
- Heeney JL, Dalgleish AG, Weiss RA (2006) Origins of HIV and the evolution of resistance to AIDS. Science 313: 462-466.
- http://www.who.int/hiv/data/en/
- Hutchinson AB, Farnham PG, Dean HD, Ekwueme DU, del Rio C, et al. (2006) The economic burden of HIV in the United States in the era of highly active antiretroviral therapy: evidence of continuing racial and ethnic differences. J Acquir Immune Defic Syndr 43: 451-457.
- Mora C, Tittensor DP, Adl S, Simpson AG, Worm B (2011) How many species are there on Earth and in the ocean? PLoS Biol 9: e1001127.
- Cleaveland S, Haydon DT, Taylor L (2007) Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol 315: 85-111.
- Pulliam JR, Dushoff J (2009) Ability to replicate in the cytoplasm predicts zoonotic transmission of livestock viruses. J Infect Dis 199: 565-568.
- Holmes EC, Drummond AJ (2007) The evolutionary genetics of viral emergence. Curr Top Immunol 315: 51-66.
- Schrag SJ, Weiner P (1995) Emerging infectious disease: what are the relative roles of ecology and evolution? Trends Ecol Evol 10: 319-324.
- Woolhouse ME, Gowtage-Sequeria S (2005) Host range and emerging and reemerging pathogens. Emerg Infect Dis 11: 1842-1847.
- Woolhouse ME, Haydon DT, Antia R (2005) Emerging pathogens: the epidemiology and evolution of species jumps. Trends in Ecol Evol 20: 238-244.
- Wolfe ND, Daszak P, Kilpatrick AM, Burke DS (2005) Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg Infect Dis 11: 1822-1827.
- Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, et al. (2005) Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci U S A 102: 7994-7999.
- Githeko AK, Lindsay SW, Confalonieri UE, Patz JA (2000) Climate change and vector-borne diseases: a regional analysis. Bull World Health Organ 78: 1136-1147.
- Daszak P, Cunningham AA, Hyatt AD (2001) Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop 78: 103-116.
- Daszak P, Cunningham AA, Hyatt AD (2000) Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science 287: 443-449.
- Paweska JT, Blumberg LH, Liebenberg C, Hewlett RH, Grobbelaar AA, et al. (2006) Fatal human infection with rabies-related Duvenhage virus, South Africa. Emerg Infect Dis 12: 1965-1967.
- Haydon DT, Cleaveland S, Taylor LH, Laurenson MK (2002) Identifying reservoirs of infection: a conceptual and practical challenge. Emerg Infect Dis 8: 1468-1473.
- Wolfe ND, Prosser TA, Carr JK, Tamoufe U, Mpoudi-Ngole E, et al. (2004) Exposure to nonhuman primates in rural Cameroon. Emerg Infect Dis 10: 2094-2099.
- Childs JE, Richt JA, Mackenzie JS (2007) Introduction: conceptualizing and partitioning the emergence process of zoonotic viruses from wildlife to humans. Curr Top Microbiol Immunol 315: 1-31.
- Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, et al. (2007) Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 13: 1031-1037.
- Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, et al. (2010) Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc Natl Acad Sci U S A 107: 16262-16267.
- Rimoin AW, Kisalu N, Kebela-Ilunga B, Mukaba T, Wright LL, et al. (2007) Endemic human monkeypox, Democratic Republic of Congo, 2001-2004. Emerg Infect Dis 13: 934-937.
- Moya A, Holmes EC, Gonzalez-Candelas F (2004) The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol 2: 279-288.
- Holmes EC (2006) The evolution of viral emergence. Proc Natl Acad Sci U S A 103: 4803-4804.
- Leendertz FH, Lankester F, Guislain P, Néel C, Drori O, et al. (2006) Anthrax in Western and Central African great apes. Am J Primatol 68: 928-933.
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, et al. (2005) Fruit bats as reservoirs of Ebola virus. Nature 438: 575-576.
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, et al. (2005) Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Taï Forest, Côte d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol 79: 12515-12527.
- Halpin K, Young PL, Field HE, Mackenzie JS (2000) Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol 81: 1927-1932.
- Yob JM, Field H, Rashdi AM, Morrissy C, van der Heide B, et al. (2001) Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis 7: 439-441.
- Towner JS, Pourrut X, Albariño CG, Nkogue CN, Bird BH, et al. (2007) Marburg virus infection detected in a common African bat. PLoS One 2: e764.
- Li W, Shi Z, Yu M, Ren W, Smith C, et al. (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679.
- Badrane H, Tordo N (2001) Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders. J Virol 75: 8096-8104.
- Rouquet P, Froment JM, Bermejo M, Kilbourn A, Karesh W, et al. (2005) Wild animal mortality monitoring and human Ebola outbreaks, Gabon and Republic of Congo, 2001-2003. Emerg Infect Dis 11:283-290.
- Wang LF, Shi Z, Zhang S, Field H, Daszak P, et al. (2006) Review of bats and SARS. Emerg Infect Dis 12: 1834-1840.
- Murray K, Selleck P, Hooper P, Hyatt A, Gould A, et al. (1995) A morbillivirus that caused fatal disease in horses and humans. Science 268: 94-97.
- Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PS, et al. (1999) Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354: 1257-1259.
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, et al. (2000) Nipah virus: a recently emergent deadly paramyxovirus. Science 288: 1432-1435.
- Luby SP, Gurley ES, Hossain MJ (2009) Transmission of human infection with Nipah virus. Clin Infect Dis 49: 1743-1748.
- Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, et al. (2009) Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001-2007. Emerg Infect Dis 15: 1229-1235.
- Plowright RK, Foley P, Field HE, Dobson AP, Foley JE, et al. (2011) Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.). Proc Biol Sci 278: 3703-3712.
- Duke-Sylvester SM, Bolzoni L, Real LA (2011) Strong seasonality produces spatial asynchrony in the outbreak of infectious diseases. J R Soc Interface 8: 817-825.
- Knobel DL, Cleaveland S, Coleman PG, Fèvre EM, Meltzer MI, et al. (2005) Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ 83: 360-368.
- Childs JE, Curns AT, Dey ME, Real AL, Rupprecht CE, et al. (2001) Rabies epizootics among raccoons vary along a North-South gradient in the Eastern United States. Vector Borne and Zoonotic Dis 1: 253-267.
- Flamand A, Coulon P, Lafay F, Kappeler A, Artois M, et al. (1992) Eradication of rabies in Europe. Nature 360: 115-116.
- Velasco-Villa A, Reeder SA, Orciari LA, Yager PA, Franka R, et al. (2008) Enzootic rabies elimination from dogs and reemergence in wild terrestrial carnivores, United States. Emerg Infect Dis 14: 1849-1854.
- Daoust PY, Wandeler AI, Casey GA (1996) Cluster of rabies cases of probable bat origin among red foxes in Prince Edward Island, Canada. J Wildl Dis 32: 403-406.
- Leslie MJ, Messenger S, Rohde RE, Smith J, Cheshier R, et al. (2006) Bat-associated rabies virus in Skunks. Emerg Infect Dis 12: 1274-1277.
- http://www.cdc.gov/lyme/stats/index.html
- Salkeld DJ, Lane RS (2010) Community ecology and disease risk: lizards, squirrels, and the Lyme disease spirochete in California, USA. Ecology 91: 293-298.
- Gubler DJ (1998) Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11: 480-496.
- Mousson L, Dauga C, Garrigues T, Schaffner F, Vazeille M, et al. (2005) Phylogeography of Aedes (Stegomyia) aegypti (L.) and Aedes (Stegomyia) albopictus (Skuse) (Diptera: Culicidae) based on mitochondrial DNA variations. Genet Res 86: 1-11.
- http://whqlibdoc.who.int/publications/2009/9789241547871_eng.pdf
- Diallo M, Sall AA, Moncayo AC, Ba Y, Fernandez Z, et al. (2005) Potential role of sylvatic and domestic African mosquito species in dengue emergence. Am J Trop Med Hyg 73: 445-449.
- Gould EA, de Lamballerie X, Zanotto PM, Holmes EC (2003) Origins, evolution, coadaptations within the genus Flavivirus. Adv Virus Res 59: 277-314.
- Meroz D, Yoon SW, Ducatez MF, Fabrizio TP, Webby RJ, et al. (2011) Putative amino acid determinants of the emergence of the 2009 influenza A (H1N1) virus in the human population. Proc Natl Acad Sci U S A 108: 13522-13527.
- Alter G, Heckerman D, Schneidewind A, et al. (2011) HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476: 96-100.
- Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, et al. (2005) Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 102: 10652-10657.
- Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, et al. (2006) Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog 2: e7.
- Bellini WJ, Harcourt BH, Bowden N, Rota PA (2005) Nipah virus: an emergent paramyxovirus causing severe encephalitis in humans. J Neuroviro 11: 481-487.
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562.
- Lindenbach BD, Thiel H-J, Rice CM (2007) Flaviviridae: The Viruses and Their Replication. Field's Virology (5th edition Volume 2). Lippincott Williams & Wilkins, Philadelphia, PA, USA.
- Schmit VL, Patton TG, Gilmore RD Jr (2011) Analysis of Borrelia burgdorferi Surface Proteins as Determinants in Establishing Host Cell Interactions. Front Microbiol 2: 141.
- Pepin KM, Lass S, Pulliam JR, Read AF, Lloyd-Smith JO (2010) Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat Rev Microbiol 8: 802-813.
- Holmes EC (2003) Error thresholds and the constraints to RNA virus evolution. Trends Microbiol 11: 543-546.
- Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, et al. (2010) Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329: 676-679.
- Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, et al. (2010) Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328: 1529.
- http://www.cdc.gov/h1n1flu/estimates_2009_h1n1.htm
- http://www.who.int/csr/disease/avian_influenza/country/cases_table_2011_08_09/en/index.html
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, et al. (2009) Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathogens 5: e1000536.
- Taylor DJ, Leach RW, Bruenn J (2010) Filoviruses are ancient and integrated into mammalian genomes. BMC Evol Biol 10: 193.
- Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, et al. (2006) Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 80: 7481-7490.
- Zhu HC, Chu DK, Liu W, Dong BQ, Zhang SY, et al. (2009) Detection of diverse astroviruses from bats in China. J Gen Virol 90: 883-887.
- Chu DK, Poon LL, Guan Y, Peiris JS (2008) Novel astroviruses in insectivorous bats. J Virol 82: 9107-9114.
- Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, et al. (2010) Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467: 420-425.
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, et al. (2006) RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biology 4: e3.
- http://www.cdc.gov/hiv/topics/surveillance/
- Mallewa M, Fooks AR, Banda D, Chikungwa P, Mankhambo L, et al. (2007) Rabies encephalitis in malaria-endemic area, Malawi, Africa. Emerging Infectious Diseases 13: 136-139.
- Mebatsion T, Cox JH, Frost JW (1992) Isolation and characterization of 115 street rabies virus isolates from Ethiopia by using monoclonal antibodies: identification of 2 isolates as Mokola and Lagos bat viruses. J Infect Dis 166: 972-977.
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. (2007) The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25: 5086-5096.
- Baric RS (2008) SARS-CoV: lessons for global health. Virus Research 133:1-3.
Relevant Topics
- Behavioral epidemiology
- Cancer epidemiology
- Disambiguation
- Economic epidemiology
- Emerging Infection
- Environmental epidemiology
- Epidemiology and Biostatistics
- Epidemiology and community health
- Epidemiology and disease control
- Epidemiology and infection
- Epidemiology of tuberculosis
- Etiology
- Genetic epidemiology
- Global Health
- HIV surveillance
- Intestinal epidemiology
- Nutrition epidemiology
- Oral/dental epidemiology
- Pediatric epidemiology
- Primary care epidemiology
- Renal epidemiology
- Reproductive Epidemiology
- Trends in maternal mortality
- Veterinary epidemiology
Recommended Journals
Article Tools
Article Usage
- Total views: 15010
- [From(publication date):
January-2012 - Jul 04, 2025] - Breakdown by view type
- HTML page views : 10293
- PDF downloads : 4717