Research Article Open Access
Vibrio Related Diseases in Aquaculture and Development of Rapid and Accurate Identification Methods
Shruti Chatterjee1* and Soumya Haldar2
1National Institute of Oceanography, Regional Centre, Lokhandwala Road, Four Bungalows, Andheri (West), Mumbai, India
2Discipline of Marine Biotechnology and Ecology, Central Salt and Marine Chemicals Research Institute (CSIR), GB Marg, Bhavnagar, Gujarat, India
- *Corresponding Author:
- Shruti Chatterjee
Scientist Fellow (QHS), Biological Oceanography Division
National Institute of Oceanography, Regional Center
Lokhandwala Road, Four Bungalows
Andheri (West), Mumbai- 400053, India
Tel: 0832-2450-441
Fax: 0832-2450-660
E-mail: schatterjee@nio.org
Received date: September 21, 2011; Accepted date: April 21, 2012; Published date: April 23, 2012
Citation: Chatterjee S, Haldar S (2012) Vibrio Related Diseases in Aquaculture and Development of Rapid and Accurate Identification Methods. J Marine Sci Res Dev S1:002. doi:10.4172/2155-9910.S1-002
Copyright: © 2012 Chatterjee S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Keywords
Vibrios; Molecular techniques; PCR; 16S rRNA; FISH; AFLP; RAPD
Abbreviations
PCR: Polymer Chain Reaction; AFLP: Amplified Fragment Length Polymorphism; FISH: Fluorescence In Situ Hybridization; RAPD: Random Amplified Polymorphic DNA; rep- PCR: Repetitive extragenic palindrome–PCR; RFLP: Restriction Fragment Length Polymorphism; VBNC: Viable But Non Culturable; FISHCH: FISH Following Cultivation
Introduction
Aquaculture remains a growing, vibrant and important production sector for high animal protein food that is easily digestible and of high biological value. Globally, marine and inland capture fisheries provide two-thirds of the total food fish supply and the remaining one-third being derived from aquaculture [1]. The reported global production of food fish from aquaculture, including fin fishes, crustaceans, molluscs and other aquatic animals for human consumption, reached a staggering height of 52.5 million tons in 2008. The contribution of aquaculture to the total production of capture fisheries and aquaculture continued to grow, rising from 34.5% in 2006 to 36.9% in 2008. In the period 1970-2008, the production of food fish from aquaculture increased at an average annual rate of 8.3% according to The state of World fisheries and aquaculture 2010 [2].
However, major setback in aquaculture is the sudden outbreak of diseases, especially caused by Vibrio spp. which is considered as a significant problem to the development of the sector with severe economic losses worldwide. Global estimation of disease losses, made by the World Bank in 1997, was the range of approximately US$ 3 billion per year [3]. Vibrio harveyi, Vibrio anguillarum are most frequently isolated marine Vibrio species [4-7], have been associated with large-scale losses of larval and juvenile penaeids [8] and also causing several opportunistic diseases to fishes [9-13]. Due to the plasticity of Vibrio genomes, with frequent horizontal gene transfer events, species boundaries are very narrow in the marine environment [14]. Hence, the identification of Vibrio related species isolated from the marine environment is sometimes tricky. An array of phenotypic and genomic techniques has become available for the identification of vibrios in the last three decades [15-22]. Accurate identification is the basic step for developing appropriate prophylactic measures in any aquaculture settings. In the present study a detailed review was undertaken to study on the diseases caused by vibrios in aquaculture settings and some of the important modern techniques to identify the disease causing organisms accurately.
Vibriosis in Aquaculture
Vibrios are gram-negative, ubiquitous in marine, estuarine ecosystem as well as aquaculture farm and one of the major microbiota of these ecosystems. Many vibrios are serious pathogens for animals reared in aquaculture [23-27]. Vibriosis is a most prevalent disease in fishes and other aquaculture reared organisms caused by Vibrio spp. and widely responsible for mortality for cultured aquaculture organisms worldwide [28,29]. Major Vibrio sp. viz. V. harveyi, V. parahaemolyticus, V. alginolyticus, V. anguillarum, V. vulnificus, and V. splendidus are usually associated with shrimp diseases. V. harveyi is associated with luminescent vibriosis in shrimps e.g., Litopenaeus vannamei and Penaeus monodon and it is most important etiological agent for mass mortality in P. monodon [28,30-32]. It has been reported that V. anguillarum, V. salmonicida, and V. vulnificus are among the main bacterial pathogens in several fish species [26]. The mode of infection in fish mainly consists of penetration of bacterium to the host tissue mainly by the chemotactic activity, followed by deployment of iron sequestering system and eventually damages the fish by means of extracellular products i.e. haemolysin and protease. In a recent study, mucus secretion, blood clots were reported to be common symptoms for moribund seabream (Sparus aurata) isolated from a hatchery located in Malta [33]. Details investigation concluded the infection might be due to haemolysin activity of V. harveyi infection. Some of the common symptoms of disease in fish caused by strains of pathogenic vibrios include intestinal nacrosis, anemia, ascetic fluid, petechial hemorriages in the muscle wall, liquid in the air bladder etc. It has been reported that shrimps are also infected by vibrios and the possible route of infection are feed, gill, hepatopancreas etc. Vibrios colonized the host tissue of shrimps after crossing the epithelial cells [34]. Some of the important aquaculture diseases caused by different Vibrio sp. are described in (Table 1). Although marine vibrios were reported to be main cause for bacterial disease in aquaculture, an important challenge for understanding the virulence potential of marine Vibrio and its mechanism for causing disease in a simple and reliable animal model is lacking. Recently, the brine shrimp, Artemia nauplii has been used in many studies mainly because of its simple culture method in gnotobiotic condition [35]. Bacterial interaction or colonization with challenged organisms is a very complex mechanism. To study the colonization potentials, several techniques have been employed such as use of cell lines [36], direct observation with scanning electron microscopy etc. [37]. These processes involve fixation and preparation of samples for microscopy, but do not permit observations in-vivo or on recently killed organisms, or guarantee that the observed bacteria are those inoculated or of interest. Thus, in-vivo colonization potential after labeling a pathogenic Vibrio strain with fluorescent dye was considered to be an appropriate method to study the colonization potential in-vivo. In a recent study Haldar et al. [38] have established pathogenic potential of a handful numbers of V. campbellii isolates, marine bacterial species, which was previously considered to be a nonpathogenic Vibrio [38].
| Vibrio spp. caused disease | Host organism | Disease | PCR based diagnostic method | Reference |
|---|---|---|---|---|
| Vibrio harveyi V. alginolyticus V. parahaemolyticus V. anguillarum V. vulnificus V. ordalii V. salmonicida Moritella viscosa (Vibrio viscosus) | Peneause monodon (Tiger prawn) Litopenaeus vannamei (White shrimp) Epinephelus coioides (Grouper) Sulculus diversicolor (Japanese abalone) P. monodon (Tiger prawn) P. monodon (Tiger prawn) Salmo salar L. (Salmon), Oncorhynchus mykiss (Rainbow trout) Oreochromis niloticus (Nile tilapia), Eels Salmonids Atlantic salmon, cod Atlantic salmon, cod |
Luminescent vibriosis resulting in mass mortality Up to 85% mortality in nauplii Gastroenteritis followed by mass mortality Mass mortality Shell disease Red disease, upto 80% mortality Vibriosis Vibriosis Vibriosis Vibriosis Winter ulcer |
Yes Yes Yes Yes Yes No No No | [84] [36] [85] [86] [87] [88] [88] [89] [90] [91] [90] [90] [90] [90] |
Table 1: Disease caused by Vibrio spp. in aquaculture.
Problems in Modern Aquaculture
In recent past intensive mode of culture with high stocking density become popular in different south east Asian countries like Thailand, Indonesis, Philippines etc. To maintain the productivity of such an intensive aquaculture, high input of fish protein originating from the sea have been employed for feeding, together with high level of water exchange and massive use of antibiotics. The spread of antibiotic resistance from aquaculture settings to natural environment is increasing. About 70% of the Vibrio isolated from aquaculture settings in Mexico are multi drug resistant [39]. On the other hand some of the important negative environmental impacts include loss of wild fishes (5 kg of wild fish has to be caught to feed 1 kg of carnivorous fish reared), loss of natural habitat, effluent discharge and destruction of sensitive habitat [40,41]. Ben-Haim et al. [41] in 2003 have advanced the hypothesis that aquaculture settings serve as foci or reservoir for pathogenic Vibrio strains, during certain period of the year, pathogenic Vibrio would withstand environmental conditions within aquaculture settings and when favourable environmental conditions established, Vibrio would be able to cause disease in wild animal [42].
Due to increasing trend of antibiotic resistance in aquaculture many alternative methods were in use by aquaculture scientist in recent time to reduce Vibrio related diseases. Among many others, some of the popular methods were use of probiotic, immunestimulants.
Different Methods Used for Bacterial Identifications
An array of molecular techniques is gaining popularity now-adays for the identification of different aquaculture related bacterial pathogens. SuiTable genetic fingerprinting methods are essential for rapid and accurate tracking of different marine vibrios. Among DNA sequence based identification, analysis of 16S rRNA and other housekeeping gene sequencing are most popular and precise methods use now-a-days to identify closely related Vibrio. Among other methods, ribotyping and PCR-based techniques, e.g., Amplified Fragment Length Polymorphism (AFLP), Fluorescence In Situ Hybridization (FISH), Random Amplified Polymorphic DNA (RAPD), repetitive extragenic palindrome (rep) –PCR (rep-PCR), and Restriction Fragment Length Polymorphism (RFLP) have also yielded the most valuable information about and new insights into the identification of closely related marine bacteria. Below we have discussed some of these methods commonly used for identification:
PCR based identification
Although there is a handful numbers of methods for identification of marine Vibrio as described above but majority of them require two or more step approaches like PCR and sequencing (16S rRNA and MLST) PCR and digestion with restriction enzymes (PCR-RFLP, AFLP) or use of radio isotope labelled probe which is expensive, time consuming and also comparatively hazardous for health. Simple and rapid identification method of Vibrio causing disease to aquaculture settings is essential for taking preventive and curative measures in aquaculture. PCR-based identification is a suiTable alternative because it is comparatively easy, less expensive and can be completed within several hours [43]. However, success of this method depends on the selection of target gene, which should be species-specific, widely distributed and also sTable in the genome.
Maximum works have been reported to identify V. harveyi related marine bacteria using PCR. Because V. harveyi is the major causal organism of luminous vibriosis, which causes potential devastation to diverse ranges of marine invertebrates over a wide geographical area. These microorganisms, however, are extremely difficult to identify because they are phenotypically diverse. Recently in 2006, Bramhachari and Dubey [44] developed PCR based identification method for V. harveyi targeting partial 16S rRNA gene [44]. Further Fukui and Sawabe [45] have modified the method by developing one step colony PCR targeting same 16S rRNA gene to identify pathogenic V. harveyi from aquaculture settings [45]. Similarly, Conejero and Hydryda [46] in 2003 have targeted toxR gene for identification of V. harveyi from aquaculture system [46]. However, most precise method to identify V. harveyi along with V. campbellii and V. parahaemolyticus was developed by Haldar et al. [47] in 2010, using multiplex PCR. This method was so accurate that the individual detection limit of all three-target species ranged from 10 to 100 cells per PCR tube, using primer concentration of 0.25 to 0.5 μmol/l (Figure 2a and 2b) [47]. Details of PCR primers for identification of some important marine Vibrio were mentioned in (Table 2). Photobacterium damselaessp. Damselae and Photobacterium damselae ssp. Piscida are important aquaculture pathogens which are responsible for causing fish disease, photobacteriosis, also known as pasteurellosis or pseudotuberculosis. High mortalities of P. piscicida infection were first observed in natural population of white perch (Morone americanus) and striped bass (Morone saxatalis) [48,49]. Photobacterium damselae ssp. which was formerly classified as V. damsela is a halophilic bacterium causing skin ulcers in warm and cold water fish [50-53]. In 2003, Rajan et al. [54] have developed a common PCR based method to identify both P. damselae ssp. Damselae and P. damselae ssp. Piscida targeting capsular polysaccharide gene and further differentiate both the species by selective culturing in on thiosulphate citrate bile salts–sucrose agar (TCBS-1) [54]. P. damselae ssp. Damselae grew on TCBS-1 producing green colonies whereas P. damselae ssp. Piscicida did not grow.
| Primer | Sequence (5'-3') | Target species | Expected bandsize (bp) | Acc. Number | References |
|---|---|---|---|---|---|
| Vca-hly5 | CTATTGGTGGAACGCAC | V. campbellii | 328 | AB271112 | [73] |
| vca-hly3 | GTATTCTGTCCATACAAAC | ||||
| Vh-hly 1F | GAGTTCGGTTTCTTTCAAG | V. harveyi | 454 | DQ224369 | [73] |
| Vh-hly 1R | TGTAGTTTTTCGCTAATTTC | ||||
| Vp-tlh1 | GATTTGGCGAACGAGAAC | V. parahaemolyticus | 695 | M36437 | [73] |
| Vp-tlh2 | CGTCTCGAACAAGGCG | ||||
| VctoxR403F | GAAGCTGCTCATGACATC | V. cholerae | 275 | CP000627 | [69] |
| VctoxR678R | AAGATCAGGGTGGTTATTC | ||||
| WhA870F | ACTCAACTATCGTGCACG | V. vulnificus | 366 | AB124802 | [69] |
| WhA1236R | ACACTGTTCGACTGTGAG | ||||
| Vng F2 | CCCGAACGAAGCGAAA | V. nigripulchritudo | 258 | [92] | |
| VngR2 | ACCTTTCAGTGGCAAGATG | ||||
| CPS F | AGGGGATCCGATTATTACTG | Photobacterium damselae | 410 | AB074290 | [80] |
| CPS R | TCCCATTGAGAAGATTTGAT | sp. Piscicida | |||
| F-gyrB | ATTGAGAACCCGACAGAAGCGAAG | V. alginoloticus | 340 | AF007288 | [93] |
| R-gyrB | CCTAATGCGGTGATCAGTGTTACT | ||||
| HG-F1 | GCTCTGTCGGAAAACTTGA | Grimontia hollisae | 363 | AB027462 | [94 |
| HG-R1 | ATGCTCAAAATGGAACACAG | ||||
| Vc_dnaJF1 | CGGTTCGYGGTGTTTCAAAA | V. coralliilyticus | 128 | [95] |
Table 2: PCR primers for identification of some important marine Vibrio.
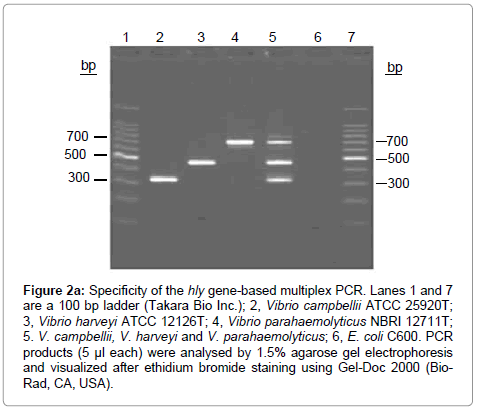
Figure 2a: Specificity of the hly gene-based multiplex PCR. Lanes 1 and 7 are a 100 bp ladder (Takara Bio Inc.); 2, Vibrio campbellii ATCC 25920T; 3, Vibrio harveyi ATCC 12126T; 4, Vibrio parahaemolyticus NBRI 12711T; 5. V. campbellii, V. harveyi and V. parahaemolyticus; 6, E. coli C600. PCR products (5 μl each) were analysed by 1.5% agarose gel electrophoresis and visualized after ethidium bromide staining using Gel-Doc 2000 (Bio- Rad, CA, USA).
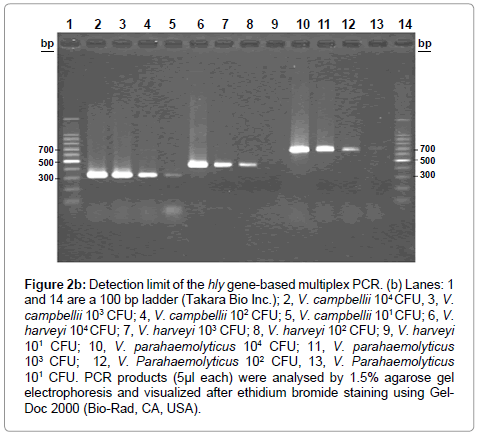
Figure 2b: Detection limit of the hly gene-based multiplex PCR. (b) Lanes: 1 and 14 are a 100 bp ladder (Takara Bio Inc.); 2, V. campbellii 104 CFU, 3, V. campbellii 103 CFU; 4, V. campbellii 102 CFU; 5, V. campbellii 101 CFU; 6, V. harveyi 104 CFU; 7, V. harveyi 103 CFU; 8, V. harveyi 102 CFU; 9, V. harveyi 101 CFU; 10, V. parahaemolyticus 104 CFU; 11, V. parahaemolyticus 103 CFU; 12, V. Parahaemolyticus 102 CFU, 13, V. Parahaemolyticus 101 CFU. PCR products (5μl each) were analysed by 1.5% agarose gel electrophoresis and visualized after ethidium bromide staining using Gel- Doc 2000 (Bio-Rad, CA, USA).
16S rRNA and housekeeping gene based identification
16S rRNA gene sequencing is considered to be very reliable method for identification of any bacteria including marine Vibrio by many authors [55-60]. The 16S rRNA gene (about 1,500 bp in length) consists of highly conserved regions and present in almost all bacteria which may reveal deep-branching (e.g., classes, phyla) relationships but variable regions may also be demonstrated which can discriminate species within the same genus. This feature has prompted researchers to use 16S rRNA both as a phylogenetic marker and as an identification tool [61].
Colony hybridization by species-specific probes
It has been demonstrated that different selective media are not quite selective or species-specific. Detection of different marine bacteria on selective media and subsequent colony hybridization with species-specific probes (probe is a fragment of DNA or RNA of variable length, which is used in DNA or RNA samples to detect the presence of nucleotide sequences), based on variable target regions of the 16S rRNA gene and other specific gene has been evaluated as a fast screening alternative tool for identification of different marine bacteria [62-66]. However, there is nearly 100% 16S rRNA gene homology among many closely related bacterial species, viz. V. scophthalmi and V. ichthyoenteri and thus there is a great possibility of cross-hybridization and misidentification of closely related species [64].
Fluorescent in situ hybridization
Fluorescent In Situ Hybridization provides a powerful tool for identifying the location of cloned DNA sequence by using fluorescence probe binds to that part of chromosomes with which they show higher degree of similarity and often used in the field of microbial ecology [67]. It has been reported that certain bacteria are metabolically active however may not be able to grow on the selective media e.g., V. cholera. Nutrient limitation or starvation, variation of pH, salinity and temperature could lead to such stage, for which they proposed the name “viable but non culturable” (VBNC) [56,68]. VBNC form of marine bacteria can be identified by direct extraction of nucleic acids from environmental samples (e.g. water, tissue, sediment etc.), followed by clonal library and 16S rRNA sequencing or alternative FISH of filter fixed cells with oligonucleotide probes targeting the 16S rRNA and subsequent visualization by epifluorescent microscopy. The low fluorescence intensity of marine bacteria is one of the main drawbacks of FISH technology [69,70]. On the other hand, because several Vibrio species (e.g. V. harveyi, V. campbellii, V. rotiferianus, and other closely phylogenetic neighbours) have very similar 16S rRNA sequences, it may be difficult to perform reliable species identification. Recently, one-step multi probe FISH method has been developed. In short, FISH method has combined with microcolony formation culture and known as FISH following cultivation (FISHFC). It has the advantage of increasing not only the specificity of probes but also the development of microcolonies in selective media within a short time which increases its applicability. A probe-reacted to the microcolonies and generate stronger fluorescence signals rather than for single colony [71]. In FISHFC variety of group or species-specific probe can be used.
Ribotyping
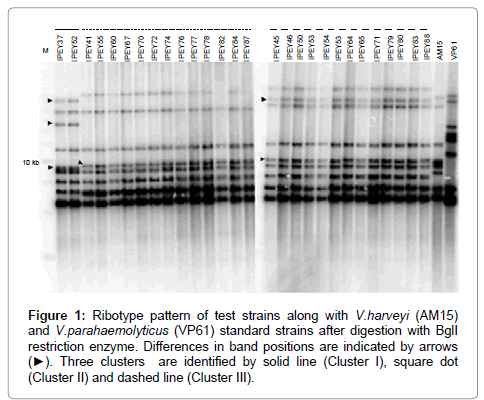
Ribotyping was one of the first fingerprinting techniques to be successfully used in the taxonomy of vibrios, and it has been particularly useful in the study of V. cholerae [72,73]. This technique is mainly used for epidemiological purposes. More recently, ribotyping has been used to assess the genomic diversity of environmental Vibrio strains associated with fish and oysters [74]. According to Austin et al. [75], closely related Vibrio species, e.g., V. anguillarum and V. ordalii, can be differentiated on the basis of ribotyping [75]. In another recent study Haldar et al. [38] have successfully differentiate two very closely related Vibrio species such as V. harveyi and V. campbellii using ribotyping method (Figure 1). Ribotyping consists of four main steps: (i) restriction of the bacterial chromosome with an endonuclease, (ii) gel electrophoresis of the resulting fragments, (iii) transfer of the fragments to a membrane, and (iv) hybridization of the gel with a labelled probe complementary to the 16S and 23S rRNAs [72]. This method is very sensitive but comparatively lengthy.
Figure 1: Ribotype pattern of test strains along with V.harveyi (AM15) and V.parahaemolyticus (VP61) standard strains after digestion with BglI restriction enzyme. Differences in band positions are indicated by arrows (âÂ?º). Three clusters are identified by solid line (Cluster I), square dot (Cluster II) and dashed line (Cluster III).
Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism or RFLP is a technique that exploits variations in homologous DNA sequences. It refers to a difference between samples of homologous DNA molecules that come from differing locations of restriction enzyme sites, and to a related laboratory technique by which these segments can be illustrated. In RFLP analysis, the DNA sample is broken into pieces (digested) by restriction enzymes and the resulting restriction fragments are separated according to their lengths by gel electrophoresis. Simple and rapid RFLP method was developed by Saha et al. [76] in 2006, based on chromosomal ori sequence of V. cholerae. It was effective to delineate between two closely related biotypes of pathogenic Vibrio strains. In a recent study, Chowdhury et al. [77], has developed an RFLP method targeting some parts of super integron region of V cholerae. genome showed good delineation between different biotype of V. cholerae strains.
Amplified Fragment Length Polymorphism (AFLP)
The AFLP is another successful PCR assay based method for differentiating the closely related Vibrio species. The AFLP method consists of mainly three steps: (i) digestion of total genomic DNA with two restriction enzymes and subsequent ligation of the restriction half-site-specific adaptors to all restriction fragments; (ii) selective amplification of these fragments with two PCR primers that have corresponding adaptor and restriction site sequences as their target sites; and (iii) electrophoretic separation of the PCR products on polyacrylamide gels with selective detection of fragments which contain the fluorescently labeled primer and computer-assisted numerical analysis of the band patterns [55]. Actually radioactively labeled primers were originally used in 1995 by Vos et al. [78] but fluorescent based primers used for performing AFLP now.
Random Amplified Polymorphic DNA (RAPD)
RAPD is a rapid, powerful and inexpensive PCR method, using arbitrary primers to detect the segment of DNA in genome. No knowledge for the DNA sequence of the targeted gene is required, as the primers will bind somewhere in the sequence is not certain. In recent years, RAPD has been used to characterize, and trace the phylogeny of diverse plant and animal species. In V. harveyi it has been used to differentiate pathogenic and non-pathogenic strain and having the ability of making different cluster whereas in other Vibrios for diversity study [79-81]. Some of the main drawback of this method includes PCR is an enzymatic reaction, therefore the quality and concentration of template DNA, concentrations of PCR components, and the PCR cycling conditions may greatly influence the outcome. Thus, the RAPD technique is notoriously laboratory dependent and needs carefully developed laboratory protocols to be reproducible.
Conclusion
In the present study all the bacterial identification were based on culturing the strain in suiTable media, followed by extraction of DNA and final identification based on the diversity of DNA sequences. No emphasis was given to identify non culturable Vibrio which comprises major percentage of total Vibrio population except in FISH. The rapid development of molecular biological techniques offers significant advantages for workers involved in fish disease diagnosis. Using nucleic acid as targets, and new methods of analysing polymorphism in these nucleic acids, can improve specificity, sensitivity and speed of diagnosis and offer means of examining the relationships between genotype and phenotype of various pathogens. Further investigation and explanation of the biodiversity among marine Vibrios by using new molecular techniques will be an important topic for future research. In recent years, the number of new publications describing new molecular techniques or methods has increased significantly. Such publications describe the development of new methods that appear very promising and useful. However, reports of application of these techniques on a routine basis in diagnostic laboratories are few. In order for molecular biology to fulfil the promise of improved diagnosis and to be adopted by regulatory authorities, thorough trials of new methods are required and the results of these must be disseminated. Molecular methods have slowly established a place in the diagnosis of disease in aquaculture.
Acknowledgements
We are thankful to Dr S R Shetye Director NIO, Dr S. N. Gajbhiye, chief scientist, NIO and Dr. N. Ramaiah, chief scientist, NIO for extending permission, providing facilities and encouragement to complete this review article. This is NIO contribution number 5076.
References
- FAO corporate document repository (2003) The role of aquaculture in improving food security and nutrition.
- http://www.fao.org/docrep/013/i1820e/i1820e.pdf
- Subasinghe RP, Bondad-Reantaso MG, McGladdery SE (2001) Aquaculture development, health and wealth. In: Aquaculture in the Third Millennium. Technical Proceedings of the Conference on Aquaculture in the Third Millennium ed., Subasinghe RP, Bueno P, Phillips MJ, Hough C, McGladdery SE, et al., (Eds.) Rome, Italy: NACA, Bangkok and FAO, 167-191.
- Arias CR, Macian MC, Aznar R, Garay E, Pujalte MJ (1999) Low incidence of Vibrio vulnificus among Vibrio isolates from seawater and shellfish of the western Mediterranean coast. J Appl Microbiol 86: 125-134.
- Pujalte MJ, Ortigosa M, Macian MC, Garay E (1999) Aerobic and facultative anaerobic heterotrophic bacteria associated to Mediterranean oysters and seawater. Int Microbiol 2: 259-266.
- Pujalte MJ, Sitjà-Bobadilla A, Macián MC, Belloch C, Alvarez-Pellitero P, et al. (2003) Virulence and molecular typing of Vibrio harveyi strains isolated from cultured dentex, gilthead sea bream and European sea bass. Syst Appl Microbiol 26: 284-292.
- Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, et al. (2011) Vibrio anguillarum as a ?sh pathogen: virulence factors, diagnosis and prevention. J Fish Dis 34: 643-661.
- Diggles BK, Moss GA, Carson J, Anderson CD (2000) Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Dis Aquatic Org 43: 127-137.
- Hispano C, Nebra Y, Blanch AR (1997) Isolation of Vibrio harveyi from an ocular lesion in the short sunfish (Mola mola). Bull Eur Ass Fish Pathol 17: 104-107.
- Company R, Sitja-Bobadilla A, Pujalte MJ, Garay E, Alvarez-Pellitero P, et al. (1999) Bacterial and parasitic pathogens in cultured common dentex, Dentex dentex L. J Fish Dis 22: 299-309.
- Alcaide E, Gil-Sanz C, Sanjuan E, Esteve D, Amaro C, et al. (2001) Vibrio harveyi causes disease in seahorse, Hippocampus sp. J Fish Dis 24: 311-313.
- Liu PC, Lin JY, Chuang WH, Lee KK (2003) Infectious gastroenteritis caused by Vibrio harveyi (V. carchariae) in cultured red drum, Sciaenops ocellatus. J Appl Ichthyol 19: 59-61.
- Zorrilla I, Morin˜igo MA, Castro D, Balebona MC, Borrego JJ (2003) Intra specific characterization of Vibrio alginolyticus isolates recovered from cultured fish in Spain. J Appl Microbiol 95: 1106-1116.
- Fraser C, Hanage WP, Spratt BG (2007) Recombination and the nature of bacterial speciation. Science 26: 476-480.
- Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, et al. (1996) Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60: 407-438.
- Rademaker JLW, Louws FJ, de Brujin FJ (1998) Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting: 1-27. In:Van Elsas JD et al. (ed.), Molecular microbial ecology manual, vol. 3.4.3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Savelkoul PH, Aarts HJ, de Haas J, Dijkshoorn L, Duim B, et al. (1999) Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol 37: 3083-3091.
- Olive DM, Bean P (1999) Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol 37: 1661-1669.
- Rademaker JL, Hoste B, Louws FJ, Kersters K, Swings J, et al. (2000) Comparison of AFLP and rep-PCR genomic fingerprinting with DNA- DNA homology studies: Xanthomonasas a model system. Int J Syst Evol Microbiol 50: 665-677.
- Dijkshoorn L, Towner KJ, Struelens M (2001) New approaches for the generation and analysis of microbial typing data. Elsevier, Amsterdam, The Netherlands.
- Gurtler V, Mayall BC (2001) Genomic approaches to typing, taxonomy and evolution of bacterial isolates. Int J Syst Evol Microbiol 51: 3-16.
- van Belkum A, Struelens M, de Visser A, Verbrugh H, Tibayrenc M (2001) Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin Microbiol Rev 14: 547-560.
- Austin B, Austin DA (1993) Vibriosis, Bacterial fish pathogens and disease in farmed wild fish. Chapter 13, Ellis Harwood Ltd. England 263-287.
- Hjeltnes B, Roberts RJ (1993) Vibriosis, In Bacterial Diseases of Fish, pg. 109-121. Edited by Inglis V, Roberts RJ, Bromage NR. Oxford: Blackwell Scientific.
- Lightner DV (1993) Diseases of cultured penaeid shrimp. In CRC Handbook of Mariculture, 2nd edn, 1: 393-486. Edited by McVey JP. Boca Raton, FL: CRC Press.
- Austin B, Austin DA (ed.) (1999) Bacterial fish pathogens. Disease of farmed and wild fish. Springer/Praxis Publishing, Chichester, UK.
- Bergh O, Nilsen F, Samuelsen OB (2001) Diseases, prophylaxis and treatment of the Atlantic halibut Hippoglossus hippoglossus: a review. Dis Aquat Org 48: 57-74.
- Lavilla-Pitogo CR, Leano EM, Paner MG (1998) Mortalities of pond-cultured juvenile shrimp Penaeus monodon associated with dominance of luminescent vibrios in the rearing environment. Aquaculture 164: 337-349.
- Chen FR, Liu PC, Lee KK (2000) Lethal attribute of serine protease secreted by Vibrio alginolyticus strains in Kurama Prawn Penaeus japonicus. Zool Naturforsch 55: 94-99.
- Lavilla-Pitogo CR, de la Pena LD (1998) Bacterial diseases in shrimp (Penaeus monodon) culture in Philippines. Fish Pathol 33: 405-411.
- Austin B, Pride AC, Rhodie GA (2003) Association of a bacteriophage with virulence in Vibrio harveyi. J Fish Dis 26: 55-58
- Guzm´an GA, Mart´inez JGS R, Casta˜neda P, Monz´on AP, Rodr´iguez TT, et al. (2010) Pathogenicity and Infection Route of Vibrio parahaemolyticusin American White Shrimp, Litopenaeus vannamei. J World Aqua Soc41: 464-470
- Haldar S, Maharajan A, Chatterjee S, Hunter SA, Chowdhury N, et al. (2010a) Identification of Vibrio harveyi as a causative bacterium for a tail rot disease of sea bream Sparus aurata from research hatchery in Malta. Microbiol Res 165: 639-648.
- Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, et al. (2001) DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol 67: 2354-2359.
- Austin B, Austin D, Southerland R, Thompson F, Swings J (2005) Pathogenicity of vibrios to rainbow traut (Oncothynchus mykiss, Walbaum) and Artemia nauplii. Environ Microbiol 7: 1488-1495.
- Olsson JC, Westerdahl A, Conway PL, Kjelleberg S (1992) Intestinal colonization potential (Scophtalmus maximis) and dab (Limanda limanda)-associated bacteria with inhibitory effects against Vibrio anguillarum. Appl Environ Microbiol 58: 551-556.
- Lavilla-Pitogo CR, Baticados MCL, Cruz-Lacierda ES, de la Pena LD (1990) Occurrence of luminous bacterial disease of Penaeus monodon nauplii in Philippines. Aquaculture91: 1-13
- Haldar S, Chatterjee S, Sugimoto N, Das S, Chowdhury N, et al. (2011) Identification of Vibrio campbellii isolated from diseased farm-shrimps from south India and establishment of its pathogenic potential in an Artemia model. Microbiology157: 179-188.
- Molina-Aja A, Garcia-Gasca A, Abreu-Grobois A, Bolan-Mejia C, Roque A, et al. (2002) Plasmid profiling and antibiotic resistance of Vibrio strains isolated from cultured penaeid shrimp. FEMS Microbiol Lett 213: 7-12.
- Kautsky N, Lubchenco J, Primavera J, Williams M (1998) Nature’s subsidies to shrimp and salmon farming. Nature 282: 883-884.
- Naylor RL, Goldburg RJ, Primavera JH, Kautsky N, Beveridge MC, et al. (2000) Effect of aquaculture on world fish supplies. Nature 405: 1017-1024.
- Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, et al. (2003) Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53: 309-315.
- Neogi SB, Chowdhury N, Asakura M, Hinenoya A, Haldar S, et al. (2010) A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett Appl Microbiol 51: 293-300.
- Bramhachari PV, Dubey SK (2006) Rapid and specific detection of luminous and non-luminous Vibrio harveyi isolates by PCR amplification. Curr Sci 90: 1105-1108.
- Fukui Y, Sawabe T (2007) Improved one step colony PCR for detection of Vibrio harveyi. Microbes Environ 22: 1-10.
- Conejero MJU, Hedreyda CT (2003) Isolation of partial toxR gene of Vibrio harveyi and design of toxR-targeted PCR primers for species detection. J Appl Microbiol 95: 602-611.
- Haldar S, Neogi SB, Kogure K, Chatterjee S, Chowdhury N, et al. (2010) Development of a haemolysin gene-based multiplex PCR for simultaneous detection of Vibrio campbellii,Vibrio harveyi and Vibrio parahaemolyticus.Lett Appl Microbiol 50: 146-152.
- Smith SK, Sutton DC, Fuerst JA, Reichelt JL (1991) Evaluation of the genus Listonella and reassignment of Listonella damsela (Love et al.) MacDonell and Colwell to the genus Photobacterium as Photobacterium damsela comb. nov. with an emended description. Int J Syst Bacteriol 41: 529-534.
- Truper HG, De Clari L (1997) Taxonomic note: necessary correction of specific epithets formed as substantives (nouns) in opposition. Int J Syst Bacteriol 47: 908-909.
- Love M, Fisher DT, Hose JE, Farmer JJ, Hickman FW, et al. (1981) Vibrio damsela, a marine bacterium, causes skin ulcer on the damselfish Chromis punctipinnis. Science 214: 1139-1140.
- Sakata T, Matsuura M, Shimkawa Y (1989) Characteristics of Vibrio damsela isolated from diseased yellowtail Seriola quinqueradiata. Bull Japanese Soc Fisheries Sci 55: 135-141.
- Fouz B, Conchas RF, Magarinos B, Amaro C, Toranzo AE (1992a) Vibrio damsela strain virulence for fish and mammals. FHS/AFS Newsletter 20: 56-59.
- Fouz B, Larsen JL, Nielsen B, Barja JL, Toranzo AE (1992b) Characterization of Vibrio damsela strain isolated from turbot Scophthalmus maximus in Spain. Dis Aqua Organisms 12: 155-166.
- Rajan PR, Lin JHY, Ho MS, Yang HL (2003) Simple and rapid detection of Photobacterium damselae ssp. piscicida by a PCR technique and plating method. J Appl Microbiol 95: 1375-1380.
- Gomez-Gil B, Soto-Rodri´guez S, Garci´a-Gasca A, Roque A, VazquezJuarez R, et al. (2004) Molecular identi?cation of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 150: 1769-1777.
- Maugeri TL, Carbone M, Fera MT, Gugliandolo C (2006) Detection and differentiation of Vibrio vulnificus in seawater and plankton of a coastal zone of the Mediterranean Sea. Res Microbiol 157: 194-200.
- Chatterjee S, Haldar S, Asakura M, Yamasaki S, Balasubramanian T (2008) Molecular identification and phylogenetic status of marine Bacillus associated with coral sediment, showing antibacterial effects against human pathogens Annal Microbiol 58: 309-312.
- Gugliandolo C, Lentini V, Spano` A, Maugeri TL (2010) Conventional and molecular methods to detect bacterial pathogens in mussels. Let Appl Microbiol 52: 15-21.
- Manmadan K, Sasaki H, Haldar S, Yamasaki S, Nagata S (2006) Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. Annal Microbiol 56: 167-173.
- Haldar S, Mody KH, Jha B (2011b) Abundance, diversity and antibiotic resistance pattern of Vibrio spp. In coral ecosystem of Kurusadai island. J Basic Microbiol 51: 153-162.
- Wiik R, Stackebrandt E, Valle O, Daae FL, Rodseth OM, et al. (1995) Classification of fish-pathogenic vibrios based on comparative 16S rRNA analysis. Int J Syst Bacteriol 45: 421-428.
- Marti´nez-Picado J, Alsina M, Blanch AR, Cerda M, Jofre J (1996) Species-specific detection of Vibrio anguillarumin marine aquaculture environments by selective culture and DNA hybridization. Appl Environ Microbiol 62: 443-449.
- Cerda-Cue´llar M, Jofre J, Blanch AR (2000) A selective medium and a speci?c probe for detection of Vibrio vulni?cus. Appl Environ Microbiol 66: 855-859.
- Cerda-Cue´llar M, Blanch AR (2002) Detection and identi?cation of Vibrio scophthalmi in the intestinal microbiota of ?sh and evaluation of host speci?city. J Appl Microbiol 93: 261-268.
- Tanaka R, Ootsubo M, Sawabe T, Tajima K, Vandenberghe J, et al. (2002) Identification of Vibrio halioticoli by colony hybridization with non-radioisotope labeled genomic DNA. Fish Sci (Tokyo) 68: 227-229.
- Sloan E, O’Neill M, Kaysner C, DePaola A, Nordstrom JL, et al. (2003) Evaluation of two nonradioactive gene probes for the enumeration of Vibrio parahaemolyticusin crabmeat. J Rapid Methods Autom Microbiol 11: 297-311.
- Thompson FL, Iida T, Swings J (2004) Biodiversity of Vibrios. Microbiol Mol bio Rev 68: 403-431.
- Xu Q, Dziejman M, Mekalanos JJ (2003) Determination of the transcriptome of Vibrio choleraeduring intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA 100: 1286-1291.
- Eilers H, Pernthaler J, Glockner FO, Amann R (2000a) Culturability and In situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol 66: 3044-3051.
- Eilers H, Pernthaler J, Amann R (2000b) Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl Environ Microbiol 66: 4634-4640.
- Sawabe T, Yoshizawa A, Kawanishi Y, Komatsu-Takeda E, Nakagawa S, et al. (2009) Multi-Probe-Fluorescence in situ Hybridization for the Rapid Enumeration of Viable Vibrio parahaemolyticus. Microbes Environ 24: 259-264.
- Grimont F, Grimont PAD (1986) Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol 137B: 165-175.
- Grimont PAD, Grimont F (2001) rRNA gene restriction pattern determination (Ribotyping) and computer interpretation, pp. 107-133. In:Dijkshoorn L, Towner KJ, and Struelens M (ed.). New approaches for the generation and analysis of microbial typing data. Elsevier, Amsterdam, The Netherlands.
- Austin B, Austin DA, Blanch AR, Cerda M, Grimont PAD, et al. (1997) A comparison of methods for the typing of fish-pathogenic Vibrio spp. Syst Appl Microbiol 20: 89-101.
- Austin B, Alsina M, Austin DA, Blanch AR, Grimont PAD, et al. (1995) Identification and typing of Vibrio anguillarum: a comparison of different methods. Syst Appl Microbiol 18: 285-302.
- Saha A, Deb R, Shah S, Ramamurthy T, Shinoda S, et al. (2006) PCR-based identification of Vibrio cholerae and the closely related species Vibrio mimicus using chromosomal ori sequence of Vibrio cholerae. FEMS Microbiol Lett 257: 84-91.
- Chowdhury N, Asakura M, Neogi SB, Hinenoya A, Haldar S, et al. (2010) Development of simple and rapid PCR-fingerprinting methods for Vibrio cholerae on the basis of genetic diversity of the superintegron. J Appl Microbiol 109: 304-312.
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, et al. (1995) AFLP: a new technique for dna-?ngerprinting. Nucleic Acids Res. 23: 4407-4414.
- Somarny WMZ, Mariana NS, Neela V, Rozita R, Raha AR (2002) Differentiation of pathogenic Vibrio species by RAPD. J Med Sci 2: 165-169.
- Alavandi SV, Manoranjita V, Vijayan KK, Kalaimani N, Santiago TC (2006) Phenotypic and molecular typing of Vibrio harveyi isolates and their pathogenicity to tiger shrimp larvae. J Appl Microbiol 43: 566-570.
- Maiti B, Shekar M, Khushiramani R, Karunasagar I, Karunasagar I (2009) Evaluation of RAPD-PCR and protein profile analysis to differentiate Vibrio harveyi strains prevalent along the southwest coast of India. J Genet 88: 273-279.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 26630
- [From(publication date):
June-2013 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 20491
- PDF downloads : 6139