Editorial Open Access
Toward In Vivo Gene Transcript Targeting MRI in Addiction Research and Therapy
Philip K. Liu*Associate Professor of Radiology, Massachusetts General Hospital and Harvard Medical School, Boston MA
- *Corresponding Author:
- Philip K. Liu
Associate Professor of Radiology
Massachusetts General Hospital and Harvard Medical School
Boston MA
E-mail: philipl@nmr.mgh.harvard.edu
Received August 28, 2012; Accepted August 29, 2012; Published August 31, 2012
Citation: Liu PK (2012) Toward In Vivo Gene Transcript Targeting MRI in Addiction Research and Therapy. J Addict Res Ther S3:e001. doi:10.4172/2155-6105.S3-e001
Copyright: © 2012 Liu PK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Addiction Research & Therapy
The Challenge, Innovation, and Impact
Advances in molecular biology, genetics and live imaging, allow direct comparison between invertebrates (nematodes, fruit flies) or vertebrates (zebra fish) and human development and neurological disorders. Genetic altered reporter genes has gained some momentum for neural cell tracking, however, not all cell types in the body share similar transfection capability for reporter genes. There remains a gap between our understanding of the gene mechanism that underlines the evolution of diseases or disorders. Fluorescent gene probes have also made advances in our ability to see connectivity and functional aspects of the central nervous systems in higher species; these modalities may yet to achieve direct imaging on molecular basis of living brains because current imaging modality suffers insufficient depth of penetrance.
Recent technological developments in magnetic resonance (MR) hardware and contrast agents have enabled high resolution of the soft brain tissue in living subjects at the anatomical, functional and molecular levels. MRI is a relatively safe technology that has excellent depth of penetration [1-4]. Nanoparticle-enhanced MR imaging (MRI) is highly sensitive and it can track neural progenitor cells in vivo after stem cell therapy in preclinical and perhaps clinical disease models. MRI measures the relaxation time of proton spins in tissue water when a magnetic field (B0) is applied; the presence of a contrast agent in the tissue alters the longitudinal (T1) and transverse (T2) relaxation times, or rate of relaxivity (R1 or R2, respectively), and the resulting MR image shows the difference of these parameters. Shimming prior to data acquisition in a 9.4 Tesla MR system can temporarily reduce the potential contribution of noise from static B0 in homogeneity.
Contrast agent-assisted MRI is considered to be most sensitive with some specificity [5-7]. However, recent method to label neural cells with non-targeting iron oxide as a contrast agent for longitudinal detection by non-invasive MRI has not generated reliable and consistent results due to contrast dilution as a result of cell division [4]. The challenges are to develop neural cell imaging methods with potential applications in humans with improved senstitivy and specificity.
Addiction Research and Therapy
Addiction to drug of abuse is a chronic disorder of the brain that severely hampers personal function and the productivity of many members of our society. Children born to drug-addicted mothers may acquire certain traits that mimic addiction-related deficits, even though they themselves are not addicted. Neuroscience research on addictions to drug has been focused primarily on the morphological and molecular adaptations of neural cells in response to chronic drug exposure. Although drugs of abuse are chemically diverse, their actions in the brain converge to produce some common effects [8] and activate similar brain regions within the mesolimbic dopaminergic pathway; such regions include the nucleus accumbens (NAc), striatum, and medial pre-frontal cortex in the forebrain [9]. Similar epigenetic influences may be related to food intake [10]. Anatomically, the A10 mesolimbic reward pathway consists of neurons originating in the ventral tegmentum (VTA) that project and terminate in the NAc, dorsal striatum (caudate putamen), and medial prefrontal cortex (mPFC). The NAc shell has been linked with emotional and motivational functions, while the NAc core appears to be involved in movement-related behaviors [11,12]. The reward pathway functions in much the same way in mice as it does in humans, and studies in mouse models are leading the way to identifying the pathways involved in human addiction. However, we acknowledge that the mechanism of drug sensitization in animals and its contribution to addiction in humans has yet to be validated [13].
A key molecule involved in psycho stimulant addiction is fosB, a transcription factor belonging to one of the Fos families of proteins. The Fos families of transcription factors are involved in activator protein 1 (AP-1) complex formation via dimerization with themselves or Jun proteins. Interestingly, fosB has been shown to associate with cocaine-induced addictive behavior and psychological disorders such as stress and depressive-like behavior in mice, suggesting that addiction and mood disorders may share common mechanisms [14]. The fosB mRNA transcript is a truncated form of FosB mRNA that is produced by alternative splicing [15]. Until recently, there have been no specific antibodies for analysis of fosB except using Western blot for specific fragment of fosB antigen among several peptides known to be fosB
The gold standard in the study of neural activation is histology with antibodies against proteins that are involved in signal transduction. Even with innovative optogenetic approach, procedures to identify interconnection of neural cells in the brain, therse assays are not routinely performed for clinical purposes because the techniques available rely on the use of biopsy or autopsy samples, except in patients with late-stage brain tumor. Due to this limitation, current tools for successful addiction therapy rely only on the assessment of clinically defined symptoms, anatomical hallmarks in the brain and consultations.
Gene Transcript Targeting MRI
Imaging technologies in living mouse brains were developed to show an elevation of mRNA prior to protein increases [16]. The investigator team has demonstrated that small single-stranded DNA with antisense sequence binds intracellular mRNA in vivo [17]; the in vivo binding allow primer free reverse transcription ex vivo followed by PCR in situ [18]. The specificity of binding to mRNA also is shown by specific retention in cells that express such specific mRNA. In a recent conference (International Conference and Exhibition on Addiction Research and Therapy at Las Vegas, Nevada, August 20-22, 2012), they presented a sensitive and less invasive for early detection of gene transcripts with accuracy. This method is less invasive because it does not required autopsy samples, and the method is with specificity for longitudinal MRI and the opportunity to monitor responses to therapy, real time provision to alternative therapies even at basic science level.
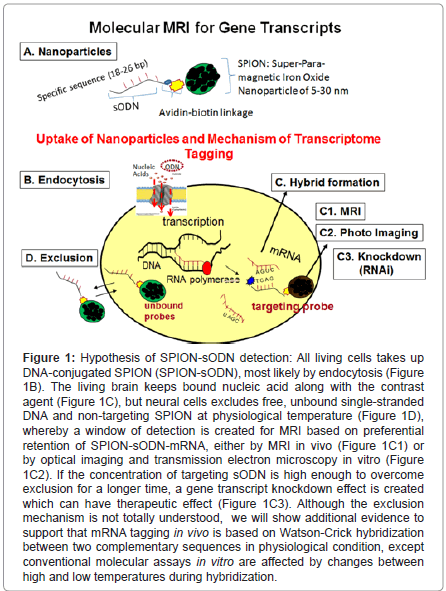
Figure 1:Hypothesis of SPION-sODN detection: All living cells takes up DNA-conjugated SPION (SPION-sODN), most likely by endocytosis (Figure 1B). The living brain keeps bound nucleic acid along with the contrast agent (Figure 1C), but neural cells excludes free, unbound single-stranded DNA and non-targeting SPION at physiological temperature (Figure 1D), whereby a window of detection is created for MRI based on preferential retention of SPION-sODN-mRNA, either by MRI in vivo (Figure 1C1) or by optical imaging and transmission electron microscopy in vitro (Figure 1C2). If the concentration of targeting sODN is high enough to overcome exclusion for a longer time, a gene transcript knockdown effect is created which can have therapeutic effect (Figure 1C3). Although the exclusion mechanism is not totally understood, we will show additional evidence to support that mRNA tagging in vivo is based on Watson-Crick hybridization between two complementary sequences in physiological condition, except conventional molecular assays in vitro are affected by changes between high and low temperatures during hybridization.
The rationale to imaging functional transcription is based on (1) gene transcripts have short half-lives, therefore represent real time gene action in life forms, (2) Watson and Crick hybridization by sequence homology can serve as the basis of mRNA tagging, (3) short DNA or RNA can be made easier than antibodies against cellular antigens and (4) antisense DNA transfects human cells. DNA-mediated transfection has been well known in literature since the early 1990s [19]. We observed the same membrane enclosed nanoparticles [20].
Translating the knowledge gained from antisense work [17,21,22], they used a very low dose of DNA (60-120 pmol DNA per kg) for MRI [23]; the dose we used is extremely small compared to the minimal dose for antisense DNA knockdown of mRNA: 10 nmol DNA per kg [17]. The gene transcript targeting MR contrast agent has three components (Figure 1A): Super paramagnetic iron oxide nanoparticles (SPION), NeutrAvidin (NA) and biotinylated phosphorothioate-modified oligoDNA (sODN), individually both iron oxide and sODN are investigational new drugs (IND). Targeting MR imaging of the brain is characterized by at least four inherent hurdles that we have to overcome: delivery, uptake, retention and exclusion of the contrast agent at an adequate concentration to produce sufficient signal over the baseline. These problems are compounded by the presence of the BBB in the central nervous system (CNS), which prohibits most macromolecules from reaching brain cells. The BBB is a formidable obstacle not only for focal macromolecular delivery, but also for global transport and distribution. Minimally invasive delivery methods to bypass the BBB include craniotomy, focused ultrasound [24,25]¸ osmotic disruption [26,27], and viral vector-assisted delivery with brain-tumor barrier disruption [28,29]. Some of these techniques hold promise in clinical application for therapeutic purposes. Cortical puncture or lumbar puncture are approved delivery routes for therapeutic drug delivery in humans (Ommaya pump), and are the only ways to guarantee focal yet global distribution in the central nervous system. Many investigators in experimental neuroscience also use cortical puncture to deliver proteins, sODN, siRNA and reporter genes [30], and optogenes [31]. This route has been used to deliver gene transcript targeting MR contrast agent in living brains by direct injection via intracerebral (icv) route (40 g Fe or 120 pmol per kg) via syringe with a 26 – 31 G needle to normal mice. In mice that already have a BBB bypass by a 26 – 31 G needle, the investigators have delivery similar agents by intraperitoneal (i.p.) injection (4 mg Fe or 12 nmol per kg) because the BBB opens for 2- 3 weeks [20,32]. In mice experienced with cerebral ischemia, this route by i.p. delivery allows a global distribution of gene transcript targeting MRI contrast agent to most brain cells [33].
This modality is highly innovative to bring molecular assays to living subjects and for cell tracking in vivo because it has several advantages:
Because proteins tends to have a longer half-lives than gene transcripts, targeting gene transcripts for detection appears to provide a real-time gene expression [33,34].
Delivery using a delivery route that is the same to that of clinically approved in humans.
The U.S. FDA has approved the sODN (single-stranded DNA with phosphorothioate modification) as IND.
The sODN we used is single-stranded DNA; therefore, it has a very minimal off-target effect by sense sequence if a double-stranded RNA (siRNA) were used.
All procedures are performed in living conditions, the modality allows longitudinal imaging/assays during the life time of the host with and without combination of neurological or behavioral tests.
Molecular assays go green: MR-contrast agent-based assays eliminate many organic solvents that need special handling [16].
The MR contrast agent tags transcripts of various genes according the sequence presented in the sODN, therefore, it has a potential to detect several gene transcript targets when various contrast agents become available as it is in gene chip application for transcriptome tagging.
Future Prospectives
If the ability to track gene activity in living subjects is realized, the impact will be tremendous for understanding disease as what really occurs in intact brains in a natural environment. The technology is not limited to identification of biomarkers in one disease mode, but could be extended to more disease models.
Acknowledgements
Research supported by NIH (1R01EB013768 & 1R01DA029889 [EUREKA award]) and Am Heart Association GIA.
References
- Modo M, Cash D, Mellodew K, Williams SC, Fraser SE, et al. (2002) Tracking transplanted stem cell migration using bifunctional, contrast agent-enhanced, magnetic resonance imaging. Neuroimage 17: 803-811.
- Ichikawa T, Högemann D, Saeki Y, Tyminski E, Terada K, et al. (2002) MRI of transgene expression: correlation to therapeutic gene expression. Neoplasia 4: 523-530.
- Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, et al. (2004) MRI detection of single particles for cellular imaging. Proc Natl Acad Sci U S A 101: 10901-10906.
- Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P (2011) Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med 65: 564-574.
- Shapiro EM, Skrtic S, Koretsky AP (2005) Sizing it up: cellular MRI using micron-sized iron oxide particles. Magn Reson Med 53: 329-338.
- Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, et al. (2003) Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology 228: 480-487.
- Bulte JW, Douglas T, Witwer B, Zhang SC, Strable E, et al. (2001) Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nat Biotechnol 19:1141-1147.
- Nestler EJ (2001) Molecular neurobiology of addiction. Am J Addict 10: 201-217.
- Paletzki RF, Myakishev MV, Polesskaya O, Orosz A, Hyman SE, et al. (2008) Inhibiting activator protein-1 activity alters cocaine-induced gene expression and potentiates sensitization. Neuroscience 152: 1040-1053.
- Wang GJ, Volkow ND, Thanos PK, Fowler JS (2004) Similarity between obesity and drug addiction as assessed by neurofunctional imaging: a concept review. J Addict Dis 23: 39-53.
- Sellings LH, Clarke PB (2003) Segregation of amphetamine reward and locomotor stimulation between nucleus accumbens medial shell and core. J Neurosci 23: 6295-6303.
- Maldonado-Irizarry CS, Kelley AE (1995) Excitotoxic lesions of the core and shell subregions of the nucleus accumbens differentially disrupt body weight regulation and motor activity in rat. Brain Res Bull 38: 551-559.
- Narendran R, Martinez D (2008) Cocaine abuse and sensitization of striatal dopamine transmission: a critical review of the preclinical and clinical imaging literature. Synapse 62: 851-869.
- Renthal W, Carle TL, Maze I, Covington HE 3rd, Truong HT, et al. (2008) Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci 28: 7344-7349.
- Nakabeppu Y, Nathans D (1991) A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell 64: 751-759.
- Liu CH, Ren JQ, Yang J, Liu CM, Mandeville JB, et al. (2009)DNA-based MRI probes for specific detection of chronic exposure to amphetamine in living brains. J Neurosci 29: 10663-10670.
- Liu PK, Salminen A, He YY, Jiang MH, Xue JJ, et al. (1994) Suppression of ischemia-induced fos expression and AP-1 activity by an antisense oligodeoxynucleotide to c-fos mRNA. Ann Neurol 36: 566-576.
- Liu CH, Huang S, Cui J, Kim YR, Farrar CT, et al. (2007)MR contrast probes that trace gene transcripts for cerebral ischemia in live animals. FASEB J 21: 3004-3015.
- Beltinger C, Saragovi HU, Smith RM, LeSauteur L, Shah N, et al. (1995) Binding, uptake, and intracellular trafficking of phosphorothioate-modified oligodeoxynucleotides. J Clin Invest 95: 1814-1823.
- Liu CH, Ren JQ, You Z, Yang J, Liu CM, et al., Noninvasive detection of neural progenitor cells in living brains by MRI. FASEB J 26: 1652-1662.
- Wahlestedt C, Golanov E, Yamamoto S, Yee F, Ericson H, et al. (1993) Antisense oligodeoxynucleotides to NMDA-R1 receptor channel protect cortical neurons from excitotoxicity and reduce focal ischaemic infarctions. Nature 363: 260-263.
- Zhang Y, Widmayer MA, Zhang B, Cui JK, Baskin DS (1999) Suppression of post-ischemic-induced fos protein expression by an antisense oligonucleotide to c-fos mRNA leads to increased tissue damage. Brain Res 832: 112-117.
- Liu CH, Kim YR, Ren JQ, Eichler F, Rosen BR, et al. (2007) Imaging cerebral gene transcripts in live animals. J Neurosci 27: 713-722.
- McDannold N, Moss M, Killiany R, Rosene DL, King RL, et al. (2003) MRI-guided focused ultrasound surgery in the brain: tests in a primate model. Magn Reson Med 49: 1188-1191.
- Mórocz IA, Hynynen K, Gudbjartsson H, Peled S, Colucci V, et al. (1998) Brain edema development after MRI-guided focused ultrasound treatment. J Magn Reson Imaging 8: 136-142.
- Rhine WD, Benaron DA, Enzmann DR, Chung C, Gonzales-Mendez R, et al. (1993) Gd-DTPA MR detection of blood-brain barrier opening in rats after hyperosmotic shock. J Comput Assist Tomogr 17: 563-566.
- Muldoon LL, Pagel MA, Kroll RA, Roman-Goldstein S, Jones RS, et al. (1999) A physiological barrier distal to the anatomic blood-brain barrier in a model of transvascular delivery. AJNR Am J Neuroradiol 20: 217-222.
- Spencer BJ, Verma IM (2007) Targeted delivery of proteins across the blood-brain barrier. Proc Natl Acad Sci U S A 104: 7594-7599.
- Rainov NG, Zimmer C, Chase M, Kramm CM, Chiocca EA, et al. (1995) Selective uptake of viral and monocrystalline particles delivered intra-arterially to experimental brain neoplasms. Hum Gene Ther 6: 1543-1552.
- Renthal W, Maze I, Krishnan V, Covington HE 3rd, Xiao G, et al. (2007) Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56: 517-529.
- Lima SQ, Miesenböck G (2005) Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121: 141-152.
- Liu CH, You Z, Ren J, Kim YR, Eikermann-Haerter K, et al. (2008) Noninvasive delivery of gene targeting probes to live brains for transcription MRI. FASEB J 22: 1193-1203.
- Liu CH, You Z, Liu CM, Kim YR, Whalen MJ, et al. (2009) Diffusion-weighted magnetic resonance imaging reversal by gene knockdown of matrix metalloproteinase-9 activities in live animal brains. J Neurosci 29: 3508-3517.
- Liu PK, Liu CH (2011) Gene targeting MRI: nucleic acid-based imaging and applications. Methods Mol Biol 711: 363-377.
Relevant Topics
- Addiction Recovery
- Alcohol Addiction Treatment
- Alcohol Rehabilitation
- Amphetamine Addiction
- Amphetamine-Related Disorders
- Cocaine Addiction
- Cocaine-Related Disorders
- Computer Addiction Research
- Drug Addiction Treatment
- Drug Rehabilitation
- Facts About Alcoholism
- Food Addiction Research
- Heroin Addiction Treatment
- Holistic Addiction Treatment
- Hospital-Addiction Syndrome
- Morphine Addiction
- Munchausen Syndrome
- Neonatal Abstinence Syndrome
- Nutritional Suitability
- Opioid-Related Disorders
- Relapse prevention
- Substance-Related Disorders
Recommended Journals
Article Tools
Article Usage
- Total views: 14833
- [From(publication date):
August-2012 - Dec 07, 2025] - Breakdown by view type
- HTML page views : 10072
- PDF downloads : 4761