Research Article Open Access
Stable Isotope Probing - A Tool for Assessing the Potential Activity and Stability of Hydrocarbonoclastic Communities in Contaminated Seawater
| Petra J Sheppard1*, Eric M Adetutu1, Alexandra Young2, Mike Manefield3, Paul D Morrison1 and Andrew S Ball1 | |
| 1RMIT University, School of Applied Science, Bundoora, Victoria 3083, Australia | |
| 2Flinders University, School of Biological Sciences, Bedford Park, SA, 5042, Australia | |
| 3Centre for Marine Bio innovation, University of New South Wales, Sydney, New South Wales, 2052, Australia | |
| Corresponding Author : | Petra J Sheppard RMIT University School of Applied Science, Bundoora Victoria 3083, Australia Tel: (61) 3 9925 6678 E-mail: Petra.Sheppard@rmit.edu.au |
| Received June 04, 2013; Accepted July 16, 2013; Published July 18, 2013 | |
| Citation: Sheppard PJ, Adetutu EM, Young A, Manefield M, Morrison PD, et al. (2013) Stable Isotope Probing - A Tool for Assessing the Potential Activity and Stability of Hydrocarbonoclastic Communities in Contaminated Seawater. J Bioremed Biodeg 4:192. doi:10.4172/2155-6199.1000192 | |
| Copyright: © 2013 Sheppard PJ, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
To optimize bioremediation strategies, knowledge of which bacterial groups are actually degrading specific hydrocarbon fractions is required. In this study, we monitored the utilization rate of unlabeled ( 12 C) and labeled ( 13 C) substrates, benzene (0.559 μL L -1 h -1 ) and hexadecane (0.330 μL L -1 h -1 ) in seawater pre-exposed to hydrocarbons over 72 h in laboratory based microcosms. Microbial community analysis by RNA-SIP DGGE showed substantial differences between the banding pattern of 12 C and 13 C communities. Cluster analysis of the microbial community profiles showed that the labeled bacterial population was ~25% similar to the original community in the unlabeled microcosms. This suggested that only a subset of the original bacterial community appeared to have utilized the labeled substrates. Sequence analysis of 16S rRNA gene sequences revealed the presence of known hydrocarbon degraders including Alcanivorax , Acinetobacter , Pseudomonas and Roseobacter . The presences of a number of Firmicutes in both sets of mesocosms suggest that these species were able to utilize both benzene and hexadecane. This study highlights the benefits of incorporating RNA-SIP in remediation studies to enhance the understanding of microbial communities in contaminated seawater.
| Keywords |
| Stable Isotope probing; Hexadecane; Benzene; DGGE; 16S rRNA |
| Introduction |
| Marine environments have a rich biodiversity which is essential for a functioning and stable ecosystem [1]. However, anthropogenic stressors such as habitat disturbances through drilling expeditions and hydrocarbon contamination threaten the biodiversity and their resilience to natural perturbations. Fortunately, microorganisms naturally play a role in carbon cycling and some have the capacity to degrade the hydrocarbons using them as a carbon and energy source [2]. Previously, more than 200 bacterial, algal and fungal genera have been identified as being able to metabolize hydrocarbons [3], indicating the ubiquity of hydrocarbon degrading capacity in the environment. Consequently, many investigations have been carried out on natural and enhanced microbial degradation of contaminant hydrocarbons in the marine environments [4,5]. |
| A fundamental question that exists while assessing and characterizing any contaminated ecosystem is identifying which types of microorganisms are present. Assessment of the microbial communities by methods such as clone library’ construction, genetic fingerprinting including denaturant gradient gel electrophoresis (DGGE), Temperature Gradient Gel Electrophoresis (TGGE), Terminal Restriction Fragment Length Polymorphism (T-RFLP) and metagenomics are approaches aimed to identify and survey the diversity of microbial communities’ [6]. Whilst metagenomics (including metatranscriptomics) can provide greater information on microbial taxonomy and metabolism than other methods and can also be used for screening based on the expression of a selected phenotype [6], these methods do not provide direct information on how individual microorganisms relate to ecosystem functioning (e.g. hydrocarbon utilization). This ability of relating specific hydrocarbon degrading activities to specific groups in the microbial community can be difficult due to the high diversity and abundance of bacterial species in the marine environment. Therefore, one major challenge when studying microbial communities is that while there is abundant information on the diversity and potential function of hydrocarbon degrading marine microorganisms, there are gaps in our knowledge with regards to which hydrocarbon fractions are actually degraded by specific microbial groups. In order to overcome this limitation Stable Isotope Probing (SIP) can be used. |
| The incorporation of stable carbon isotopes in scientific experiments has shown promising results especially in studies for validating intrinsic bioremediation [7-9], examining contaminant migration and distribution [10] and those focusing on natural isotope abundance [11]. The use of SIP in natural abundance studies has demonstrated the application of this methodology to work in situ, which is advantageous for bioremediation studies. One of the advantages of using SIP in bioremediation studies is the ability to use this technique to target specific substrates and therefore specific subpopulations. Microbial utilization of substrates such as labeled carbon 13C or nitrogen 15N results in their incorporation into cellular biomarkers such as nucleic acids (e.g. RNA, DNA) [12]. Nucleic acids which contain the labeled substrate will be heavier than nucleic acids with unlabeled fractions. These different fractions can be separated by ultracentrifugation and fractionated into aliquots by displacing the gradient with water [12]. Furthermore, molecular identification of specifically labeled populations within the ‘heavy’ stable isotope enriched nucleic acids can be identified (through sequencing), corresponding to the microorganisms which were able to metabolise the labeled substrate of interest [13]. The use of RNA-SIP holds significant potential for exploring active populations as the use of RNA achieve higher synthesis rates and requires reduced incubation times when compared to DNA-SIP [13]. |
| The use of SIP has the potential to help in clarifying the identities of key microorganisms involved in degrading contaminants during bioremediation. SIP can aid in determining whether the hydrocarbon degrading microorganisms present during bioremediation are actually involved in contaminant degradation or merely a fraction of the microbial community that can tolerate the contaminant. In most investigations on microbial roles in bioremediation, the presence of hydrocarbonoclastic organisms is often associated with hydrocarbon degrading activities (especially when substantial degradation has occurred). This assumption is usually made with little or no information on the proportion of microbial groups that were directly involved in biodegradation. In addition there is limited information on microbial community dynamics during the bioremediation of a contaminated marine environment. Questions remain on which microbial groups are directly involved in the process, what phases of the degradation process are these groups active and the level of functional redundancy that exists within the community. |
| The aim of this study was to identify and compare the microbial communities of pre-exposed hydrocarbon contaminated seawater in closed marine systems using an RNA-SIP technique. In particular, this study focused on i) investigating and identifying the dominant microbial groups in hydrocarbon degradation through 16S rRNA analysis ii) determining if those prevalent microbial groups are actively involved in hydrocarbon mineralization using SIP, incorporating an aliphatic (hexadecane) and an aromatic substrate (benzene) and iii) understanding the dynamics of the active community using microbial community based tools such as range weighted richness (Rr), functional organization (Fo) and dynamics (Dy). |
| Materials and Methods |
| Substrate utilization microcosms |
| The 13C hexadecane and 12C benzene and their 12C versions used for this study were obtained from Sigma- Aldrich (USA) with the 13C labeled fractions having a chemical and isotope purity of 99 atom%. These hydrocarbons were selected as representatives of aromatic and aliphatic fractions commonly found in crude oil. Sterile glass serum bottles (500 mL) were each filled with seawater previously exposed (>27 weeks: Supplementary Table 1) to hydrocarbon contaminants (250 mL) (initially 1% w/v crude oil) and successfully subjected to bioremediation to completely eliminate the hydrocarbon contaminants. Prior to the start of this study, the pre-exposed seawater was subject to chemical analysis to validate the absence of hydrocarbon contaminants. |
| The microcosms were spiked with either hexadecane (13C or 12C) or benzene (13C or 12C) to a final concentration of approximately 65 μL L-1 and 30 μL L-1 respectively and immediately sealed with Tefloncoated butyl septa. The samples included i) pre-exposed seawater; ii) pre-exposed seawater with 12C substrate (unlabeled benzene UB), iii) pre-exposed seawater with 12C substrate (unlabeled hexadecane UH), iv) pre-exposed seawater with 13C substrate (labeled benzene LB), v) pre-exposed seawater with 13C labeled hexadecane LH) and vi) sterilized pre-exposed seawater (control). Replicate samples of treated and control samples were incubated over 72 h. Destructive sampling was carried out at times 0, 24, 48 and 72 h. Microcosms were incubated freestanding at room temperature with natural lighting until >40% degradation had occurred (~72 h). Autoclaved microcosms were used as controls throughout the experiment to ensure degradation was by biotic processes. All standards and controls had the same liquid/ headspace ratio and were held at the same temperature as the test samples. |
| Hydrocarbon substrate monitoring |
| Kitagawa tubes coupled with an aspirating pump AP-20 (Kitagawa, Japan) were used as per the manufacturer’s protocol to determine the substrate concentration in each benzene microcosm. Briefly, benzene concentrations were measured from the headspace of each microcosm with a Kitagawa 118SC detection tube containing a chemical reagent calibrated for measuring 1-100 ppm benzene within a temperature range of 0-40°C. Hexadecane was also analyzed using Kitagawa tubes but with mixed results as most of the substrate concentration could not be recovered. Therefore, gas chromatography was then used for hexadecane analysis using a liquid extraction method. A standard calibration curve was constructed from hexadecane concentration dilutions (hexane), and the equation from the standard calibration curve was used in conjunction with the area under each chromatogram (correlated area) to determine the hexadecane concentration in each hexadecane-treated microcosms. The hexadecane samples were analyzed by mixing hexane (5 mL) in each microcosm (triplicate) and allowing the hexadecane to dissolve in the solvent. After separation, 1 mL of the solvent was extracted from each microcosm and sampled on a HP 6890 Gas Chromatogram (GC) system equipped with a 5973 Mass Spectrometry Detector (MSD). A programmed temperature split 20:1 injection was used. The injector temperature was initially set at 60°C, and then increased at a rate of 50°C/min to 270°C, where it was maintained. The capillary column used was an Agilent JW DB5 (30 m by 0.25 mm with 0.25 μm thickness), with helium as the carrier gas flowing at a rate of 1.9 mL/min in a constant flow mode. |
| RNA extraction |
| RNA was extracted from all treatments at each of the four different time points after the preparation of the microcosms, corresponding to time 0, 24, 48 and 72 h. The RNA was extracted using a phenol-chloroform extraction method [4]. Briefly, pelleted cells were resuspended in sodium phosphate buffer (SPB, 0.5 mL, pH 7.9) and phenol-chloroform-isoamyl alcohol (0.5 mL, 25:24:1) and then lysed by bead beating for 2×20 s (Mini Bead Beater K9, BioSpec). After centrifugation (12,000×g for 5 min at 4°C) the aqueous layer was removed and purified with phenol-chloroform-isoamyl alcohol (25:24:1). The RNA was further purified, precipitated, ethanol washed and resuspended in molecular grade water (30-50 μL). DNA was digested with RNase-free DNase (Promega, USA) in accordance with the manufacturer’s protocol to remove residual DNA and purify the RNA. Test PCR was carried out using the appropriate primers to confirm the absence of DNA in the digested samples (detection of no amplicon). The extracted RNA was then subject to ultracentrifugation and fractionation. |
| Ultracentrifugation and fractionation |
| Equilibrium density gradient centrifugation and gradient fractionation were conducted in 1.99 g mL-1 cesium trifluoroacetate (GE Healthcare, UK) as per the Manefield et al. [12] protocol. Each gradient was made up of 500-800 ng of total RNA. Hexadecane samples were sealed in polyallomer bell top Quickseal centrifuge tubes (13 x 32 mm), and spun in a TLA 100.3 rotor (Beckman Coulter) in an Optima TLX Ultracentrifuge (Beckman Coulter, USA) at 120,000 rpm at 20°C for 24 h; benzene samples were sealed in polyallomer Optiseal centrifuge tubes (13 x 51 mm) and spun in a NVT90 rotor (Beckman Coulter, USA) in an Optima L-100 XP ultracentrifuge (Beckman Coulter, USA) at 45,000 rpm at 20°C for 24 h. After centrifugation, gradients were fractionated from below by displacement with water using a syringe pump at a flow rate of 3.3 μL s-1. The buoyant density of each gradient fraction was determined by weighing known volumes on a four-figure milligram balance. After the ‘density location’ of the fractions had been determined, fractions were subject to isopropanol precipitation and molecular analysis to further validate changes in the gradient density profile; separation of ‘heavy’ and ‘light’. Thereafter, only the appropriate fractions (in triplicate) (based on their density location representative of ‘heavy’ and ‘light’ [14]) were used for molecular analysis. |
| 16S rRNA amplification and denaturant gradient gel electrophoresis |
| Transcription of RNA into cDNA was performed with GoScript™ Reverse Transcriptase (Promega, USA) in accordance to the manufacturer’s protocol using reverse primer 518R (5’ATTACCGCGGCTGCTGG-3’) [15]. Two microliters of cDNA was used per 50 μL PCR mixture as previously described [16]. The primers used for PCR were 341F with GC clamp and 518R (10 pmol/μL). The amplified products were then separated on a 9% polyacrylamide gel with a 45-60% denaturant gradient. Electrophoresis was run at a constant voltage of 60 V for 18 h at 60°C in 1 X TAE running buffer. After electrophoresis, the gels were silver stained [17] and scanned with an Epson V700 scanner. Dominant bands (41, marked in Figure 1A and 1B) of interest were aseptically excised, purified and cleaned as previously described by Sheppard et al. [16]. Purified products were sent to the Australian Genome Research Facility (AGRF, Melbourne, Australia) and sequenced on an ABI Prism 3730. The nucleotide sequences were analysed using Sequencer software (Sequencer Version 5.0, Gene Codes Corporation, USA) and the consensus sequences were submitted to GenBank (see Table 2A and 2B for accession numbers). Homology searches were completed with the Basic Local Alignment Search Tool (BLAST) server of the National Centre for Biotechnology Information (NCBI) using a BLAST algorithm (http://www.ncbi.nlm. gov.library.vu.edu.au/BLAST/) for the comparison of a nucleotide query sequence against a nucleotide sequence database to determine putative identities of band sequences. |
| Data analysis |
| Digitised DGGE gel images were processed using Phoretix 1D advanced analysis package (Phoretix Ltd, UK). Banding patterns were analyzed with Phoretix to generate similarity profiles using the unweighted pair group method with mathematical averages (UPGMA) [15]. Pareto-Lorenz curves were used to estimate Functional Organization (Fo) and evenness within the microbial community [18]. Range weighted richness (Rr) were used to calculate the carrying capacity of the microbial community according to the equation Rr= (N2×Dg) where N is the total number of bands multiplied by Dg which is the percentage of denaturing gradient of the sample analyzed [18]. Dynamics (Dy) is the number of species that on average come to significant dominance which was a concept based on the moving window analysis, the rate of change (Δt) parameter [18]. For each selected profile, the percentage similarities between each time frame (T0-24 h, 24-48 h and 48-72 h) were obtained from the UPGMA dendrograms. The percentage change at each time-frame was calculated using the formula %change = 100-%similarity and (Δt) calculated as described by Marzorati et al. [18]. Statistical significance was determined using ANOVA and statistical significance accepted at P<0.05. |
| Results and Discussion |
| Substrate utilization and gradient evaluation |
| Rapid aerobic utilization of benzene was detected over a 72 h incubation period with a rate of 0.559 μL L-1 h-1 (Table 1) suggesting the presence of adapted hydrocarbon degraders. The rate of benzene utilization was higher than that observed for hexadecane (0.330 μL L-1 h-1) (P<0.05). The higher rate of benzene utilization compared to hexadecane may be a result of differences in their chemical structure and solubility properties [19]. Solubility constraints with the labeled hexadecane (partition coefficients Log Kow 8.25 and Log Kow 2.13 for hexadecane and benzene respectively) may have resulted in some quantity of the substrate not being available for microbial activity but also as a result of the different microorganisms involved. Alternatively, it may be a result of an adaptive status of the microbial communities’ given the pre exposure to weathered hydrocarbon [19]. As the samples were pre-exposed to hydrocarbon contamination; control readings taken prior to the start of the experiments confirmed that no background concentrations were present (no detectable hydrocarbon contaminant). The sterile controls showed no substantial difference in spiked hydrocarbons at the beginning and the end of the experiments (data not shown) confirming that degradation was by biotic processes. |
| After the incorporation of the labeled carbon atoms (substrate) into microbial nucleic acids as a result of biotic degradation, the RNA extracts were subjected to ultracentrifugation and a density gradient was formed (Supplementary Table 2). After validation of the separation of the ‘heavy’ and ‘light’ fractions, appropriate fractions were selected (in triplicate) at each of the four time points for both of the unlabeled and labeled samples (benzene and hexadecane). The average location of the gradient density selected corresponded to hexadecane (13×32 mm tube) labeled 1.834 (± 0.011) g mL-1, hexadecane unlabeled 1.778 (± 0.013) g mL-1, benzene (13×51 mm tube) labeled 1.798 (± 0.012) g mL-1 and benzene unlabelled 1.781 (± 0.013) g mL-1. The location densities share similar results to those seen by Manefield et al. [14]. |
| Labeled versus unlabeled fraction: bacterial community structure |
| The degradation results showed that the microbial communities contained active hydrocarbon degraders within each microcosm (Table 1). 16S rRNA gene analysis was therefore used to assess the microbial community structure and determine changes in its community composition in response to the addition of aliphatic and aromatic hydrocarbon substrates. The RNA-SIP DGGE results showed substantial differences between banding patterns from 12C and 13C bacterial communities. |
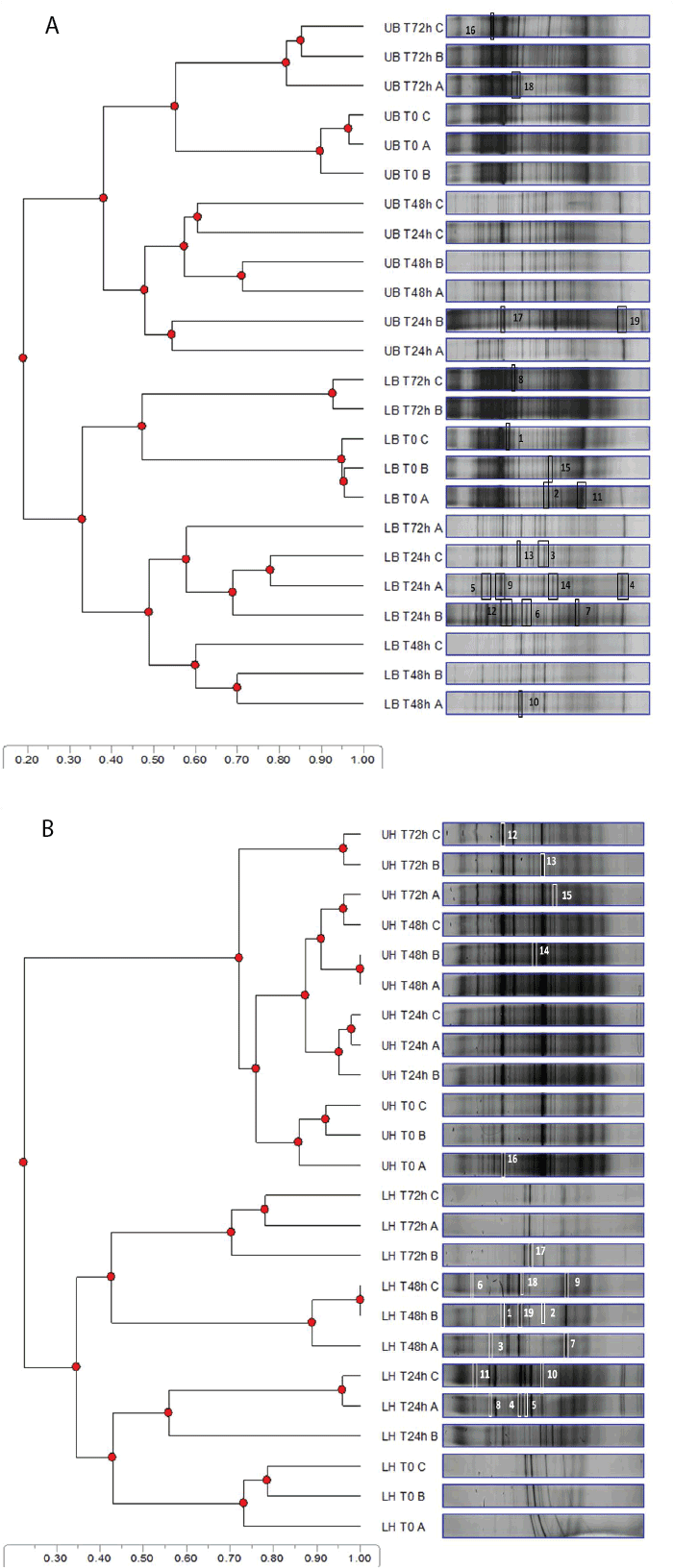
| Figure 1A shows the UPGMA dendrogram of bacterial community profiles obtained from microcosms supplemented with labeled and unlabeled benzene. The bacterial community in the unlabelled and labeled benzene microcosm each formed distinct clusters. However, there were substantial differences between the communities over the 72 h incubation period. For example, time 0-24 h communities had 0.55 and 0.48 similarity indices with 48-72 h having lower similarity indices of 0.38 and 0.34 for unlabeled and labeled microcosms respectively. There was also high replicate sample variability in some of the microcosms (Figure 1A). The microbial community in the labeled microcosms was also only ~20% similar to community in the unlabeled microcosms. |
| Figure 1B shows the UPGMA for hexadecane, which in contrast to benzene, indicates that the communities in replicate unlabeled fractions were grouped closely, ~0.73 on the Dice Sorensen’s similarity index (irrespective of sampling time). These communities were also highly similar to the no substrate control (data not shown). However, the bacterial community in the labeled hexadecane microcosm showed a similar trend to the one observed in labeled benzene microcosms. For example the community at day 0 was substantially different to that in 24 h (0.42 similarity index) and 48-72 h (0.35 similarity index) samples. The labeled fraction cluster was also only 0.23 similar to the unlabeled substrate cluster. This suggests that 20-25% (based on similarity coefficient) of the total community detected were involved in the breakdown of the benzene and hexadecane substrates at the time of sampling as well as under the investigated conditions (e.g. constant temperature). |
| However, this does not necessarily mean that this was the only fraction of the total microbial community that was capable of degrading the supplied substrate. These findings only suggest that in both benzene and hexadecane microcosm a fraction of the dominant microbial population in the unlabeled microcosms was involved in substrate utilization. This means that in the environment not all putative hydrocarbon degraders present in the community are involved in the degradation process. |
| The SIP-RNA-DGGE profile analysis also suggests that the community exposed to aromatic hydrocarbons was more sensitive than that to an aliphatic substrate. The DGGE profile was more reflective of the changes in the aromatic community (unlabeled) than the aliphatic community which had a stable community (unlabeled). Traditionally, DGGE profiles are known to reflect dominant microorganisms (top 1%) [6,15] and this dominance is assumed to have resulted from microbial activities and population. However, this study has shown that while DGGE gives a good picture of prevalent members in the community, not all the dominant microbial groups were involved in benzene and hexadecane utilization. Pre-exposure of seawater to hydrocarbon contaminants would usually have resulted in selection of hydrocarbon degrading microorganisms. The similarity cluster (Figure 1A and 1B) showed only 20-25% of this population compared to the total population in unlabeled microcosms was actually shown to have utilized the substrates. This suggests that only a subset of the dominant population in the original 12C community (assumed to be predominantly hydrocarbonoclastic) is utilizing the 13C substrate in the labeled microcosms. This could have been due the fact that the detected groups with 13C incorporated into their DNA were the only groups capable of using the supplied substrate. Alternatively, this may simply be a reflection that the communities in these microcosms were functionally redundant, rather than a lack of hydrocarbon degrading capacities in the remaining 75-80% of the population [20]. |
| In addition, the bacterial community in labeled and unlabeled benzene microcosms at 0 and 72 h (samples) were more similar to each other than to other samples collected at 24 or 48 h, unlike in hexadecane microcosms. The reason for this is not entirely clear. However, given that substantial degradation of benzene (up to 80%) had occurred by 72 h, the high similarities observed might have been due to the benzene stressed community returning to its original composition. This suggested that the bacterial community in the benzene supplemented microcosms were resilient. A key attribute of a resilient community is the ability of the altered community to return to its original composition after the substantial reduction or elimination of the stressor (benzene) [20]. By combining DGGE with RNA-SIP in this study, we were able to evaluate the dominant bacterial community and detect the fraction in this community that was actually degrading the specific hydrocarbon substrates. In the case of hexadecane, 16S rRNA-SIP was sensitive enough to pick differences which would have been undetectable using the conventional 16S rRNA based DGGE fingerprinting method. This was because the 16S rRNA-DGGE profile of the unlabeled microcosms showed very little community changes compared to the substantial changes observed in the labeled microcosm’s profile (16S rRNA-SIP) over a 72 h period (Figure 1B). |
| Reliance on the 16S rRNA-DGGE profile alone could have led to the wrong conclusion that the community was largely stable when it was a dynamic and highly changing one. Therefore changes in bacterial community during bioremediation in closed contaminated marine environments are better assessed with 16S rRNA-SIP, although experiments longer than 72 h might be needed to validate this assumption providing that problems with cross feeding are properly addressed. |
| Microbial community dynamics |
| Given the assumed adaptation of the microbial community as a result of pre-exposure to contaminated seawater, we investigated the community dynamics during the experimental period. The diversity and structure of the bacterial communities were analysed using the Pareto-Lorenz curve to assess functional organization (Fo), windows moving analysis to assess dynamics (Dy) and the range weighted richness (Rr) to assess total number of species present. |
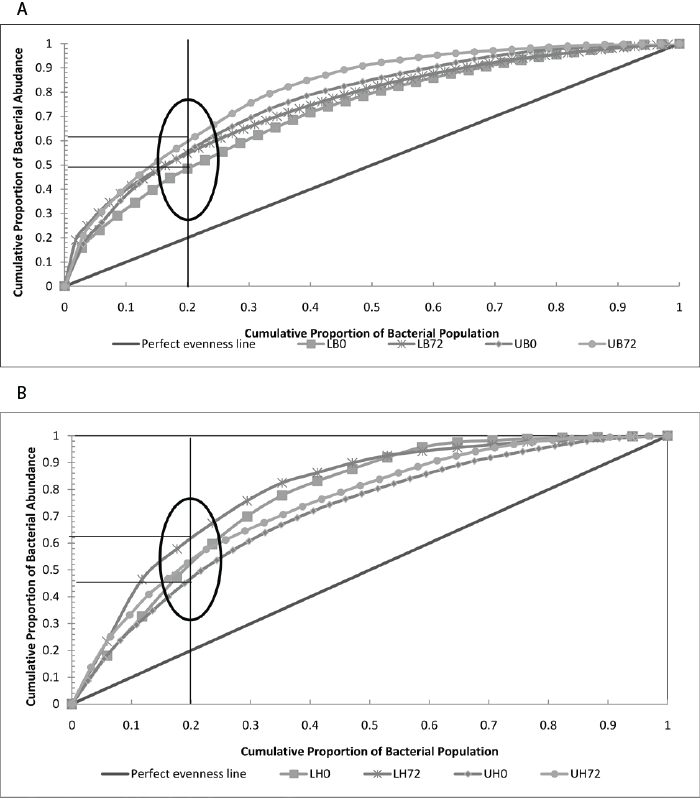
| To assess the functional organization (Fo) of the bacterial (benzene & hexadecane) community a Pareto-Lorenz curve distribution patterns were plotted of the 16S rRNA DGGE profiles. The Pareto-Lorenz curve is based on the number of bands and their intensities [21]. The functional organization of the bacterial communities in labeled and unlabeled benzene and hexadecane microcosms indicated communities with medium (45-60 %) functional organization (Figure 2A and 2B). The average functional organization indicates a community which can potentially deal with changing environmental conditions [18], such as the addition of a hydrocarbon substrate. This is logical as the samples were pre-exposed to hydrocarbon contamination which leads to adapted hydrocarbon degrading communities. Communities with medium functional organization are also assumed to be functionally redundant [18]. This appeared to be the case in this study as the UPGMA cluster analysis appears to show that only a fraction (20-25%; labeled community similarity to unlabeled community) of the total community was involved in benzene and hexadecane degradation (Figure 1). |
| The changes in a microbial community over time are reflected in the microbial community dynamics (Dy) analyses. This change over time is referred to as Δt. To evaluate the dynamics (Dy) of the 16S rRNA DGGE profiles, 24 hour-based deviations were established with moving window analysis (MWA). Δt values were very high (~62%) for the microbial communities in labeled and unlabeled benzene microcosms suggesting a high level of Dy (Supplementary Table 3). High (Dy) is reflective of a highly variable community (unstable) with broad dynamics with different species coming into dominance at different times [18]. While the Δt was also high (~59%) in labeled hexadecane communities, it was comparatively lower (~23%) in the unlabeled hexadecane community (Supplementary Table 2). This further validates the sensitivity of the RNA-SIP-DGGE methodology in picking up substantial changes in community dynamics which could have been missed by RNA-DGGE analyses which had suggested a comparatively more stable community and response to hexadecane introduction. All communities which had the incorporation of labeled benzene and hexadecane expressed high community dynamics and this was probably related to the pre-exposure (and presumably preadaptation) of the community to hydrocarbon contamination. This could have meant that members of the community had the capability to metabolize the introduced substrates and associated secondary and tertiary degradation products. |
| Similarly to Dy and Fo results, all samples showed similar results for weighted richness (Rr). The Rr value informs on the carrying capacity of an environment and whether the environment is habitable, adverse or exclusive [22]. High Rr value indicates a high microbial diversity (more bands on the DGGE profile) which correlates to an increase in the community carrying capacity and habitability. All samples produced high richness values, scoring values above 30%, therefore achieving high band numbers (diversity) on the DGGE analysis (data not shown). |
| Overall, these results indicate that all bacterial communities in the microcosms reflected active hydrocarbon degrading microorganisms. Therefore sequence analysis was performed on the microcosm communities to identify the dominant species involved in the substrate utilization in order to validate this hypothesis. |
| Sequence analysis |
| Sequence analysis of the dominant 16S rRNA bands from the DGGE profiles revealed similarities to bacterial groups known to contain hydrocarbons degrading species including affiliations to Actinobacteria, Firmicutes and Proteobacteria [23]. |
| Table 2A shows the sequence analysis for labeled benzene sequences which detected putative identities to hydrocarbon degrading species Acinetobacter, Delftia, Pseudomonas putida, Shewanella, and Stenotrophomonas [23]. Similarly, Table 2B shows the sequence analysis for labeled hexadecane microcosms. In contrast to the unlabeled and no substrate microcosm communities, the labeled hexadecane microcosms differed by the absence of the prevalent marine Arcobacter-related sequences. Additionally it differed by the presence of dominant bands whose sequences shared strong identities to known hydrocarbon degrading species including Alcanivorax, Bacillus, Brevimonas, Chryseobacterium, Pseudomonas, Rhodococcus and Roseobacter [23]. In addition sequence analysis showed similarities in the species composition with in Firmicutes. The prevalent identification of Bacillus species in both substrate microcosms is not surprising in hydrocarbon degradation studies, as some Bacillus species are able to produce biosurfactants during hydrocarbon oxidation [24] thereby reducing the interfacial tension. Therefore this study suggests that members of this group played an important role in the degradation of the hydrocarbons in the closed marine system. |
| Conclusion |
| This study has further highlighted the benefits of RNA-SIP application in linking microorganism presence to degradation, by identifying species utilizing individual substrates by the incorporation of a labeled substrate. The utilization of individual hydrocarbons by a highly active and diverse community present in pre-exposed seawater within 72 h of incubation with benzene (unlabeled and labeled) was also successfully demonstrated. Microbial community analyses suggested that only 20-25% (based on similarity values) of the original community appeared to be involved in substrate utilization. We propose that this was likely due to a functional redundant and adapted microbial community rather than a lack of capacity by the remaining members of the community for hydrocarbon contaminant removal. Furthermore, the use of SIP based fingerprinting method was shown to be more sensitive to detecting changes in the community structure than conventional fingerprinting methods. Sequence analysis of each microcosm identified putative sequences of known hydrocarbon degrading microorganisms with similarities shown between the group Firmicutes. This study therefore shows how RNA-SIP application and the use of appropriate tools for community fingerprint analysis can enhance our understanding of microbial dynamics in remediation studies in contaminated seawater. |
| Acknowledgements |
| This research was supported by the Australian India Strategic Research Fund (BFO20032). |
References
|
Tables and Figures at a glance
| Table 1 | Table 2A | Table 2B |
Figures at a glance
 |
 |
| Figure 1 | Figure 2 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 13957
- [From(publication date):
June-2013 - Oct 31, 2025] - Breakdown by view type
- HTML page views : 9316
- PDF downloads : 4641
