| Rapid Communication |
Open Access |
|
| Yash Paul Khajuria1,2*, Sanjana Kaul1 and Manoj K Dhar1 |
| 1Plant Genomics Laboratory, School of Biotechnology, University of Jammu, Jammu-180006, India |
| 2National Institute of Plant Genome Research, New Delhi-110067, India |
| *Corresponding author: |
Yash Paul Khajuria
National Institute of Plant Genome Research
New Delhi-110067, India
Tel: 09873645870
E-mail: yashkhajuria@gmail.com |
|
| |
| Received August 11, 2012; Published September 30, 2012 |
| |
| Citation: Khajuria YP, Kaul S, Dhar MK (2012) Molecular Characterization of Venturia inaequalis Causing Apple Scab in Kashmir. 1:339. doi:10.4172/scientificreports.339 |
| |
| Copyright: © 2012 Khajuria YP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. |
| |
| Abstract |
| |
| Apple scab is causing major losses to apple growers throughout the world. The disease is caused by fungal pathogen Venturia inaequalis (Cke. ) Wint., which is a heterothallic, haploid ascomycete, reproducing sexually and asexually. Understanding the genetic variability of the pathogen is very important for devising the strategies to control it. DNA based molecular markers have been successfully used for studying the genetic variation in different fungal pathogens. For the present investigation, samples were collected from the different areas of Kashmir valley, during 2004 to 2008. Two molecular markers, namely RAPD and ISSR were used for characterization of V. inaequalis isolates, obtained from different cultivars growing in various orchards spread across the valley. The gene diversity revealed by RAPD markers ranged between 0.08 and 0.64, whereas for ISSR, it ranged between 0.1 and 0.8. We conclude that there is high level of genetic diversity within populations of V. inaequalis. The isolates were observed to be intermixed and not area specific, however, in some cases, cultivar specific isolates were identified. |
| |
| Keywords |
| |
| Apple scab; Venturia inaequalis; RAPD; ISSR; Gene diversity |
| |
| Introduction |
| |
| Apple scab is one of the most serious diseases of apple and is caused by the fungus Venturia inaequalis [1]. The disease causes great economic losses to the growers in particular and apple industry, in general [2]. Although, infection may occur on leaves, fruits, stem or green twigs, the main sites of infection are leaves and fruits. Infection first appears on the underside of the leaf and subsequently, spreads to the other parts. Young leaves are more prone to infection than mature and old leaves [3]. First visible symptoms are pin head sized water soaked spots. Later, these enlarge and assume dark and smoky appearance. When the disease advances, skin ruptures and the exposed tissue gives brown or black velvet like appearance. Finally, they attain a brownish black colour. If fruits get infected before harvest, the symptoms are not visible until storage [4]. During storage, rough, black and circular lesions may develop. The lesions are formed due to erect, brown conidiophores and numerous conidia produced by the fungus. The ascospores constitute the primary source of infection. These become active as soon as spring starts [5]. The pathogen increases its biomass considerably within 7 to 10 days, and further infection takes place by asexual spores called conidia. |
| |
| In Kashmir valley, the release of the ascospores can begin as early as late March, and may continue over several weeks [6]. Further infections occur with asexual conidiospores. Conidia spread from tree to tree in rain–splash and by wind. If the weather conditions are suitable, then the fungus spreads very rapidly and there can be a dramatic increase in the incidence of scab. The pathogen remains active even at very low temperatures [7], and can infect at as low as 1°C within 40 hours. As temperature and wetness are directly related to the incidence of infection, these parameters can be used for prediction of incidence of scab [8]. A modification to Mills table has been proposed, which is relevant to Kashmir valley [9]. Even, several models for estimates of ascospore maturity have been developed for controlling the disease [10]. |
| |
| Recently established orchards have been found to be more vulnerable to scab, than those established several decades ago. This can be attributed to commercialization, whereby only one or few types of cultivars are grown in orchards. The situation can become out of control, in case of an epidemic. Therefore, it is necessary to understand the genetic diversity of the fungus so that strategies for controlling it can be devised well in time. |
| |
| In order to control the fungus, various fungicides are being used. This remains the only viable strategy to control the disease, after the infection has occurred. However, indiscriminate use of fungicides is causing serious environmental and health concerns [5,11]. Even fungicide resistance has been reported in different apple growing areas [12]. The long lasting strategy is the cultivation of scab resistant apple cultivars. Major scab resistance genes like Vf, Vbj, Vr, Vm etc., have been identified as potential sources of scab resistance in apple [12]. Among these, Vf represents the most extensively studied gene and has been introgressed in many commercial cultivars. However, the disturbing situation is the fast evolution of new races of V. inaequalis, which are overcoming scab resistance genes [13]. Therefore, study of genetic variation in V. inaequalis, using molecular markers becomes extremely important. |
| |
| DNA based markers have been used to study many pathogenic fungi [14]. RAPD is the simplest method and is generally used for preliminary studies, in unknown populations. RAPD markers have been used for characterization of V. inaequalis [15] in Czech Republic. Characterization of European population has been undertaken to understand the gene diversity, within population [16]. In this study, populations of Venturia inaequalis have been reported to be highly diverse and distributed homogeneously throughout. Kashmir valley represents the main area of cultivation of apple in India; however, no information is available on molecular characterization of Venturia. The present investigation aims at filling this lacuna, and seeks to generate information on the genetic diversity among different populations of V. inaequalis. |
| |
| Materials and Methods |
| |
| Collection of infected material |
| |
| Infected leaf samples were collected from the orchards at different time intervals, during 2003 to 2007 (Figure 1a and 1b). After collection, the leaves were placed between the folds of the newspapers for drying. Efforts were made to remove as much moisture as possible, in order to avoid cross contamination by other fungi. The samples were kept in the paper bags and were marked, indicating different accession numbers. The bags were wrapped in polythene and kept at 4°C to minimize the degradation, and to prevent the contamination. |
| |
|
|
Figure 1 (1a and 1b): Symptoms of apple scab on leaves and fruit. Note the brown or black velvet like appearance of the spots. |
|
| |
| Culturing, purification and preservation of fungal isolates |
| |
| Potato dextrose agar medium (PDA) is the basic medium for growth of many fungi. Other media tried for the culture of the fungi are apple infusion agar (AIA) and corn meal agar (CMA). The PDA medium worked very well for V. inaequalis. The antibiotic chloramphenicol (50 μg/ ml) was added to the medium, to avoid bacterial contamination. |
| |
| Leaves were treated with disinfectants to eliminate the bacteria and other non–fixed saprophytes from the surface of the leaf. Leaves were first washed with autoclaved distilled water, dipped in 70% ethanol for 10 seconds, and again washed using autoclaved distilled water. Small pieces from the infected area of the leaf were cut and washed in sterile distilled water. The leaf bits were inoculated on PDA slants and kept at 18°C. The fungal growth was observed after 10–12 days of inoculation. The isolates obtained from leaves were again sub-cultured, to obtain pure cultures. Out of several isolates obtained, finally 20 isolates were selected for further studies (Table 1). Single spores were obtained on water agar medium. For long term storage, spores were mixed in 10% glycerol in preservation vials and stored at ultra low temperature (–80°C). These cultures were stored as stock cultures for future use. |
| |
|
|
Table 1: Symptoms of apple scab on leaves and fruit. Note the brown or black velvet like appearance of the spots. |
|
| |
| Fungal DNA isolation |
| |
| Genomic DNA was extracted using the CTAB method with minor modifications [17]. After 12–15 days of inoculation, hyphal mass was obtained from flasks by filtering the broth through sterile muslin cloth. Fungal mass was then dried between the folds of sterile filter paper. It was powdered in a pre–cooled pestle and mortar, using liquid nitrogen. About 1gm of fungal powder was added to preheated 10 ml CTAB buffer in centrifuge cups, and incubated for 30 minutes at 65°C in a water bath. Samples were mixed every 10 min by inverting centrifuge cups 5–6 times, to make sure that the tissue mixed well with the buffer. 10 ml chloroform: isoamyl alcohol (25:1) was added to each tube. After proper mixing, the samples were centrifuged to sediment the cell debris and the upper aqueous phase were then transferred to another tube. Later, equal volume of chilled DNA grade ethanol was added, to precipitate DNA. Samples were mixed gently and centrifuged at 8000 rpm for 5 minutes. Supernatant was discarded and pellets were washed 3 times with 70% ethanol, air dried and dissolved in 100 μl TE. The DNA samples were treated with 1μl RNase (10 mg/ml) for three hours at 37°C, to remove RNA. |
| |
| The RNA free DNA was further purified by extracting with a mixture of phenol, chloroform and isoamyl alcohol (25:24:1), followed by centrifugation at 5000g for 10 min. The aqueous phase was collected and mixed with equal volume of chloroform, followed by centrifugation at 5000 g for 10 min. The DNA was precipitated from the aqueous phase, using chilled ethanol. The DNA was obtained as pellet after centrifugation, which was dissolved in TE and stored at –20°C. |
| |
| Quantification of DNA was done both spectrophotometrically, as well as by agarose gel electrophoresis, using lambda DNA as standard. In the latter case, band intensity of sample DNA was compared with the band intensity of lambda DNA of various concentrations, and the concentration of sample DNA was thus determined. |
| |
| RAPD (Random Amplified Polymorphic DNA) analysis |
| |
| PCR was performed in 25 μl volume containing 3 mM MgCl2, 1 × Taq polymerase buffer, 200 μM of dNTPs, 0.8 μM primer, 0.4 units of Taq polymerase and 50 ng of DNA. The thermal cycling conditions consisted of an initial denaturation for 5 minutes at 94°C, followed by 35 cycles of denaturation for 1 min at 94°C, annealing for 30s at 36°C and extension for 2 min at 72°C, with a final extension of 10 min at 72°C. |
| |
| Samples were visualized on 1.4% agarose gel. The gels were scored for the presence and absence of bands. 25 different RAPD primers were used in present study, and 8 primers showing best polymorphism were selected for further analysis. |
| |
| ISSR (Inter Simple Sequence Repeats) analysis |
| |
| Amplification was performed in a 25 μl reaction volume which contained 1 X Taq buffer, 3 mM MgCl2, 200 μM of each dNTPs, 0.5 μM of primer, 10-30 ng template DNA, and 0.6 units of Taq DNA polymerase. The thermal cycler conditions consisted of initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation for 30s at 94°C, annealing at 45°C for 30s and extension for 2 min at 72°C, with the final extension of 10 minutes. 28 ISSR primers were screened and finally 8 best primers were used for analysis. |
| |
| Results and Discussion |
| |
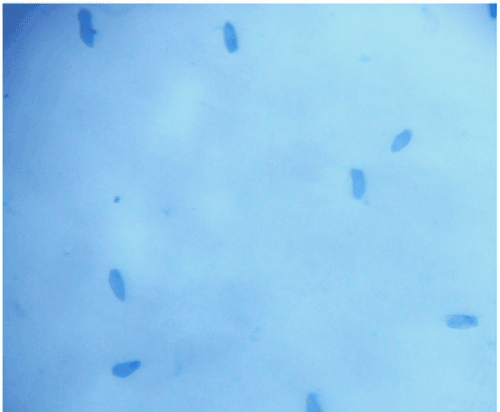
| Culturing of V. inaequalis was a difficult step because of cross contamination of other fungi. Venturia is very slow growing fungus, and takes 10–15 days for growth. Other fungi like Alternaria are very fast growing and spread on the slant within 3-4 days. We conducted detailed investigations on the morphological features of the fungus. The spores were of characteristic flame shape, in all the isolates. However, the spore size was variable; the size ranged from 13–36 x 6–12 μM (Figure 2). |
| |
|
|
Figure 2: Typical flame shaped conidiospores of Venturia inaequalis as observed under the microscope. |
|
| |
| As pointed out earlier, breeding apple cultivars with durable resistance to V. inaequalis is the need of the hour. However, to achieve this it is extremely important to understand the genetic structure of this pathogen, including its virulence elements and the extent to which evolutionary forces may influence such elements. We used two molecular markers to undertake detailed investigations on the genetic structure of Venturia. The details are as follows: |
| |
| RAPD (Random Amplified Polymorphic DNA) analysis |
| |
| |
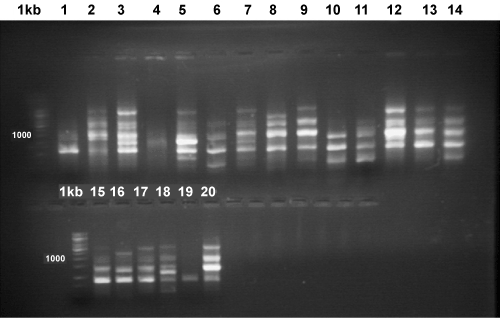
| The total numbers of fragments amplified using 8 RAPD primers were 84 (Figure 3). The primers generated 84% to 100% polymorphism. The gene diversity ranged from 0.08 to 0.64. Resolving power of primers ranged between 0.59 and 10.6. |
| |
|
|
Figure 3: RAPD profile of different isolates of Venturia after PCR amplification using random Primer OPN13. |
|
| |
| ISSR (Inter Simple Sequence Repeat) analysis |
| |
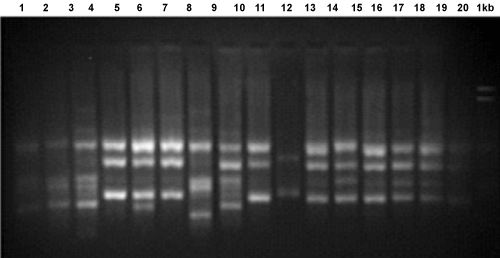
| 8 primers produced a total of 57 scorable markers, among 20 isolates. The size of amplified products ranged from 200 bp to 3400 bp. The number of scorable markers produced per primer ranged from 6 to 9 (Figure 4). All primers showed 100% polymorphism, except primer UBC 808 which showed 60% polymorphism. Gene diversity and resolving power ranged from 0.1 to 0.8 and 3.9 to 8.1, respectively. |
| |
|
|
Figure 4: ISSR profile of different isolates of Venturia after PCR amplification using primer UBC 855. |
|
| |
| Cluster analysis was performed by combining the data generated through RAPD and ISSR analyses (Figure 5). Two major clusters were obtained; cluster A and cluster B. Cluster A contains 4 isolates and cluster B contains 16 isolates. Cluster B was further divided into two sub–clusters C and D. Sub–cluster C contains maximum number of isolates, i.e. 13 and sub–cluster D contains three 3 isolates. Sub–cluster C could be further divided into two groups E and F. Group E contains 6 isolates and F contains 7 isolates. |
| |
|
|
Figure 5: Dendrogram generated by combining data of RAPD and ISSR analyses. The isolate number shown in the dendrogram is as per the serial number of the isolate given in Table 1. |
|
| |
| Using microsatellite markers on isolates of Venturia inaequalis from five continents, it has been postulated that the fungus may have originated in Central Asia, from where it has spread to other parts of the world [18]. These studies have however, revealed high within population genetic diversity in this fungus [19]. Surprisingly, though, to the best of our knowledge, there are no reports on evaluation of genetic diversity of the fungus from Kashmir valley. Since Kashmir valley is the richest repository of apple germplasm in India, therefore, it would be worthwhile to assess the genetic diversity of fungal isolates from this region. The study may help in understanding the co-evolution of the host and the pathogen especially, in this part of the world. |
| |
| The aim of the present work was to study genetic diversity of V. inaequalis from Kashmir valley. Identification of cultivar specific and area specific strains of V. inaequalis, were two parameters used for evaluation. We observed high level of genetic variation among different isolates of V. inaequalis. Several reports on molecular investigations in different fungi [20] have documented high level of variation, among isolates of a single species. In V. inaequalis, existence of high variability among different isolates has been explained on the basis of the fact that it reproduces both by sexual and asexual means [21]. In the present case, analysis of RAPD and ISSR profiles did not show any significant differences, in isolates of V. inaequalis collected from various geographical regions. This observation draws support from the dendrogram, as clustering of isolates from same area or adjacent areas was not observed. It can therefore, be concluded that these isolates are not genetically isolated from one another. |
| |
| Similar study has been conducted in Venturia populations from Switzerland, using RAPD and PCR–RFLP markers [19]. Within population diversity was found to be quite similar in populations from different countries [22]. Interestingly, the situation was found to be different in populations in United Kingdom and China at molecular level, when AFLP markers were used. While isolates obtained from different apple cultivars in the same orchard in United Kingdom differed significantly at molecular level, as well as in virulence characteristics, the same was not true of Chinese populations [22]. We observed high genetic variability within populations, but there was less difference among geographically divergent populations. Earlier reports from Europe have shown that V. inaequalis population is highly diverse and distributed homogeneously throughout the area [19]. Dissemination of spores by biotic and abiotic factors may be the cause of even distribution of genetic variation. |
| |
| Interestingly, the dendrogram analysis, in the present case, pointed towards existence of cultivar specific isolates. Isolates collected from particular cultivars tried to cluster together. For example, the isolates obtained from the Delicious cultivars clustered together. Similarly, isolates from Ambri and Hazratbali cultivars clustered together. However, more extensive studies need to be conducted, in order to reach a firm conclusion. |
| |
| The present study is first to use RAPD and ISSR markers to assess genetic diversity of V. inaequalis isolates from Kashmir valley. |
| |
| Acknowledgments |
| |
| Authors are grateful to the Department of Biotechnology (DBT), Govt. of India for funding the project on Apple. YPK is thankful to Council of Scientific and Industrial Research (CSIR) for financial assistance, in the form of Junior and Senior Research fellowships. Thanks are due to Coordinator, Bioinformatics Centre, University of Jammu for providing the necessary facilities. |
| |
| |
| References |
| |
- Bénaouf G, Parisi L (2000) Genetics of Host-Pathogen Relationships Between Venturia inaequalis Races 6 and 7 and Malus Species. Phytopathology 90: 236-242.
- Machardy WE (1996) Apple Scab: Biology, Epidemiology and Management. The American Phytopathological Society, St. Paul, MN.
- Thakur VS, Sharma RD (1999) Apple scab and its management, diseases of horticultural crops. Diseases of Horticultural Crops–Fruits, Indus Publication Co. New Delhi.
- Berrie AM, Xu XM (2003) Managing apple scab (Venturia inaequalis) and powdery mildew (Podosphaera leucotricha) using Adem. Int J Pest Manage 49: 243-249.
- Holb IJ, Heijne B, Withagen JC, Gáll JM, Jeger MJ (2005) Analysis of summer epidemic progress of apple scab at different apple production systems in the Netherlands and hungary. Phytopathology 95: 1001-1020.
- Sharma IM (2010) Antagonistic effect of fungi associated with apple scab lesions on growth of its pathogen Venturia inaequalis (Cke.) Wint. Research Journal of Agricultural Sciences 1: 245-248.
- Stensvand A, Gadoury DM, Amundsen T, Semb L, Seem RC (1997) Ascospore Release and Infection of Apple Leaves by Conidia and Ascospores of Venturia inaequalis at Low Temperatures. Phytopathology 87: 1046-1053.
- Mills WD, Laplante AA (1951) Diseases and insects in the orchard. Cornell University Agricultural Experimental Station Bulletin 711: 21-27.
- Qazi NA, Ahmad K, Beig MA, Munshi NA, Khan NA (2008) Impact of inoculum distance on the initiation and spread of apple scab infection [Venturia inaequalis (Cke.) Wint.] to isolated apple plantation. Applied Biological Research 10: 40-43.
- James J, Sutton I (1982) A model for predicting ascospores maturation of V. inaequalis. Phytopathology 72: 1081-1085.
- Pimentel D, McLaughlin L, Zepp A, Lakitan B, Kraus T, et al. (1993) Environmental and economic effects of reducing pesticide use in Agriculture. Agr Ecosyst Environ 46: 273-288.
- Jones AL, Shabi E, Ehret GR (1987) Genetics of negatively correlated cross-resistance to a N-phenylcarbamate in benomyl-resistant Venturia inaequalis. Can J Plant Pathol 9: 195-199.
- Patocchi A, Walser M, Tartarini S, Broggini GA, Gennari F, et al. (2005) Identification by genome scanning approach (GSA) of a microsatellite tightly associated with the apple scab resistance gene Vm. Genome 48: 630-636.
- Vanova M, Polisenska I, Vejl P, Skupinova S (2000) Differentiation of cereal eyespot fungi using morphologic characteristic and PCR diagnostic tests. Plant Protection Science 36: 57-64.
- Melounova M, Vejl P, Sedlak P, Reznerova A, Tesaaova M, Blazek J, Zoufala J (2004) The variability of Venturia inaequalis CKE. races in the Czech Republic and the accumulation of resistance genes in apple germplasm. Plant, Soil and Environment- UZPI 50: 416-423.
- Tenzer I, Gessler C (1997) Subdivision and genetic structure of four populations of Venturia inaequalis in Switzerland. European Journal of Plant Pathology 103: 565-571.
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13-15.
- Gladieux P, Zhang XG, Afoufa-Bastien D, Valdebenito Sanhueza RM, Sbaghi M, et al. (2008) On the origin and spread of the Scab disease of apple: out of central Asia. PLoS One 3: e1455.
- Tenzer I, Gessler C (1999) Genetic diversity of Venturia inaequalis across Europe. European Journal of Plant Pathology 105: 545-552.
- Adachi Y, Watanabe H, Tanabe K, Doke N, Nishimura S, et al. (1993) Nuclear Ribosomal DNA as a Probe for Genetic Variability in the Japanese Pear Pathotype of Alternaria alternata. Appl Environ Microbiol 59: 3197-3205.
- Gladieux P, Caffier V, Devaux M, Le Cam B (2010) Host-specific differentiation among populations of Venturia inaequalis causing scab on apple, pyracantha and loquat. Fungal Genet Biol 47: 511-521.
- Xu X, Yang J, Thakur V, Roberts A, Barbara DJ (2008) Population variation of apple scab (Venturia inaequalis) isolates from Asia and Europe. Plant Disease 92: 247-252.
|
| |
| |