Research Article Open Access
Proteomics– Current Novelties and Future Directions
Sara ten Have*, Yasmeen Ahmad and Angus I Lamond
Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, Dow Street, Dundee, DD1 5 Scotland, UK
- *Corresponding Author:
- Dr. Sara ten Have
Wellcome Trust Centre for Gene Regulation and Expression College of Life Sciences
University of Dundee, Dow Street, Dundee
DD1 5EH, Scotland, UK
Tel: +44 1382386787
E-mail: s.m.tenhave@dundee.ac.uk
Received date: August 03, 2011; Accepted date: September 07, 2011; Published date: Augutst 05, 2011
Citation: Have ST, Ahmad Y, Lamond AI (2011) Proteomics- Current Novelties and Future Directions. J Anal Bioanal Tech S3:001. doi: 10.4172/2155-9872.S3-001
Copyright: © 2011 ten Have S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Increasingly in the last decade, proteomics has become a widely used technique in the field of cell biology. This is due not only to improvements in technology and analytical procedures, but also because of the innovative and diverse ways in which the methodologies of protein and peptide analysis have been applied. This has resulted in the broadening of applications and potential uses for proteomics, including analyses of cellular substructures and mechanisms to imaging of protein distributions in cells and tissues. The growing scale and scope of modern proteomics projects has brought to the fore computing challenges involved in managing and mining the resulting very large and complex data sets. Modern proteomics is therefore inherently multidisciplinary, requiring the expertise of software engineers and statisticians as well as cell biologists, protein chemists and mass spectrometrists. This review article will describe some of the elegant, quirky, and novel applications of mass spectrometry based proteomics to medical research, diagnostics, quality control and drug discovery.
Keywords
Mass Spectrometry; Methodology; Novel applications; Proteomics
Proteomics is the term used to describe the global study of proteins in a sample. While multiple techniques can be used to detect and quantitate proteins, modern proteomics now predominantly relies upon Mass Spectrometry as the method of choice because of its high sensitivity and ability to detect proteins in high throughput. Proteomics is a relatively young science, given its current title as recently as 1994 [1], coined from the focus of the research- PROTEins and its apparent similarity with the broad ranging and high throughput analysis of genes, genOMICS.
Mass Spectrometry uses ionisation (electron loss producing a positive charge) and exact mass measurement (down to 1-2 ppm) to measure proteins and peptides (Figure 1). Typically a “bottom-up” approach is taken, where bench top chemistry and enzyme digestion is applied to protein samples extracted from cells or tissues. This generates predictable peptides and the spectra from these peptides are measured using a mass spectrometer. Additional information about the individual peptide ions can be derived in the mass spectrometer by fragmentation of the ions induced by collision with an inert gas and a further round of measurement.
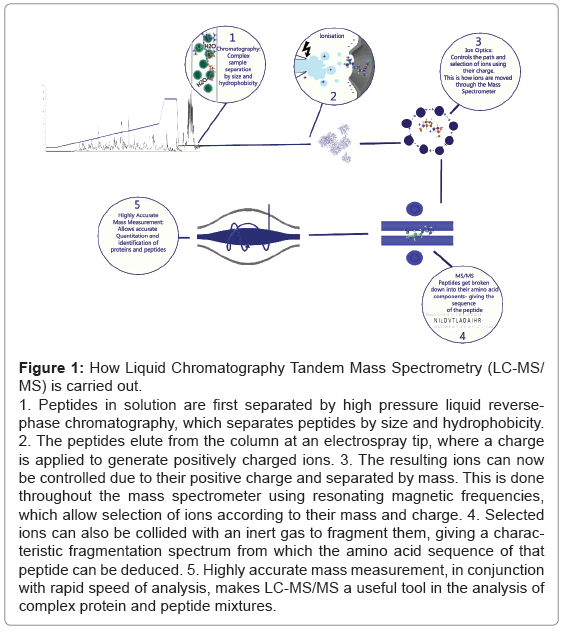
Figure 1: How Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) is carried out.
1. Peptides in solution are first separated by high pressure liquid reversephase chromatography, which separates peptides by size and hydrophobicity.
2. The peptides elute from the column at an electrospray tip, where a charge is applied to generate positively charged ions. 3. The resulting ions can now be controlled due to their positive charge and separated by mass. This is done throughout the mass spectrometer using resonating magnetic frequencies, which allow selection of ions according to their mass and charge. 4. Selected ions can also be collided with an inert gas to fragment them, giving a characteristic fragmentation spectrum from which the amino acid sequence of that peptide can be deduced. 5. Highly accurate mass measurement, in conjunction with rapid speed of analysis, makes LC-MS/MS a useful tool in the analysis of complex protein and peptide mixtures.
Because genome sequences are now available for humans and most commonly used model organisms, the resulting cell protein sequences can be predicted and an “in silico” digestion performed that creates a library of peptide masses that can be derived from these proteins. Commonly used protein databases include Uniprot for example which is species specific and hand curated, while the larger TrEMBL database is automatically annotated and not reviewed (http://www.ebi.ac.uk/ uniprot/). The exact peptide masses which are predicted based upon in silico analysis can then be compared against the actual measured masses seen in the output of the mass spectra of a sample. Matches of spectra to specific peptides can then be made, which in turn can be matched to provide information about proteins in the original sample. The analysis of spectra and matching to the predicted peptide masses relies upon dedicated software packages that can handle thousands of peptides because the scale and complexity of most samples precludes manual annotation.
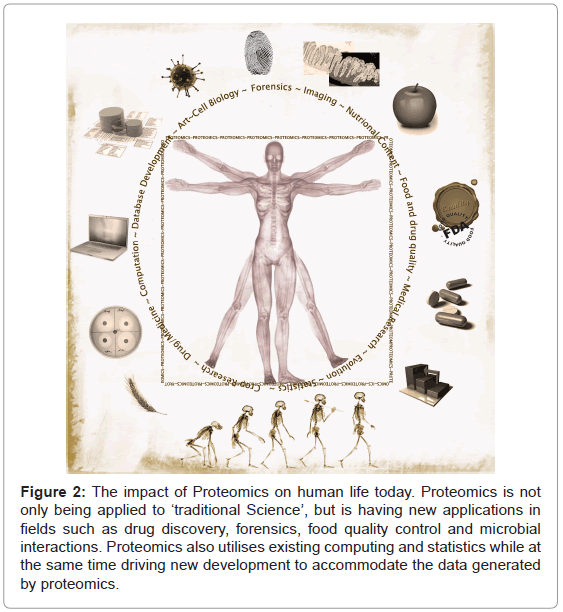
Proteomics has expanded greatly in scale and scope and is finding new and unexpected applications (Figure 2). The fundamental roles played by proteins in living systems as well as the inherently multidisciplinary nature of proteomics have led to application of this new science not only for answering basic research questions, but also in areas such as forensics, food analysis, clinical medicine and even for studying the origins of life on earth.
Figure 2: The impact of Proteomics on human life today. Proteomics is not only being applied to ‘traditional Science’, but is having new applications in fields such as drug discovery, forensics, food quality control and microbial interactions. Proteomics also utilises existing computing and statistics while at the same time driving new development to accommodate the data generated by proteomics.
The sequencing of the ‘human genome’ initially involved the sequencing of the genomes of two Caucasian males, but is now being extended rapidly to cover a much larger and ethnically representative sample of our species (for example see the Wellcome Trust Sanger Institute’s 1000 genome project (http://www.1000genomes.org/)). The current abundance of DNA sequence information has inspired genomic analysis of human disease, assuming that changes in the genetic code may be the cause of, or contribute to, the disease. This gene sequencing analysis frequently seeks to link Single Nucleotide Polymorphisms, or ‘SNPs’, with disease phenotypes. In most cases however it is likely that the cause of the disease results directly or indirectly from changes in the structure and/or expression of one or more proteins. Applying proteomics methods can thus augment genome studies and provide critical information about how the properties of cell proteins vary in different genetic backgrounds. This proteomics approach is now being applied to study cancers neurological pathologies, arthritis, cell lines from many tissue types and body fluids of every description [2,3,4,5,6]. The majority of these endeavours are aiming to discover ‘biomarkers’, i.e., characteristic changes in the structure or expression of specific proteins that can be used to construct a simple and reliable assay for the diagnosis of disease.
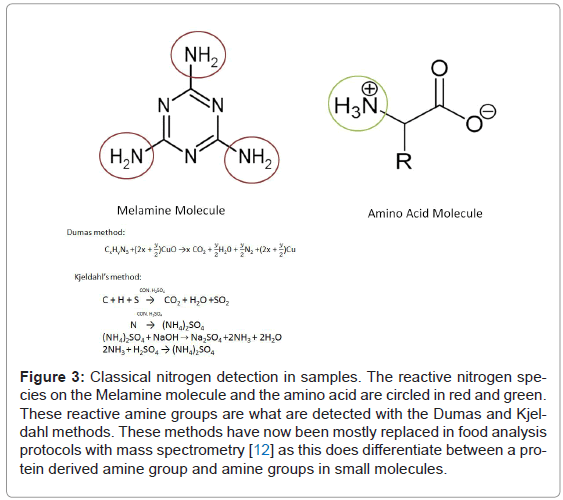
The application of proteomics to the search for biomarkers has expanded now from studying what comes out of our bodies, to include analysis of what goes into our bodies. The field of nutritional proteomics has recently proven useful in quality control of food stuffs, as well as the nutritional value. For example not only finding out what species a meat really is, as opposed to what it claims to be, but also provides information about allergen percentage in food, illegal additives and undesirable colorant addition. In 2008 we saw the not insignificant effect of unmonitored addition of melamine to dairy products in China (Figure 3). Melamine was added to artificially increase the apparent protein content of the dairy product. This resulted in ~300,000 people [7] being adversely affected by the reaction of this nitrogen boosting additive with cyanuric acid in their bodies (which forms when melamine reaches the kidneys and reacts with urine). Fifty one thousand nine hundred infants and young children were hospitalised with kidney difficulties (WHO report December 2008) because of the illegal addition of this compound. Typically dairy protein levels were determined from measuring nitrogen by analysis methods such as the Kjeldahl [8] and Dumas [9], which are widely used by production companies and quality control agencies alike. These methods use extremely high heat and either reaction with sulphuric acid to generate ammonia (which is purified and measured), or else complete combustion and measurement of purified elements (nitrogen, water and carbon dioxide) respectively. This does not differentiate between nitrogen originating from proteins and nitrogen arising from other molecules which might be present in a sample [10]. Hence the addition of nitrogen rich Melamine resulted in artificially high estimates of protein levels.
Figure 3: Classical nitrogen detection in samples. The reactive nitrogen species on the Melamine molecule and the amino acid are circled in red and green. These reactive amine groups are what are detected with the Dumas and Kjeldahl methods. These methods have now been mostly replaced in food analysis protocols with mass spectrometry [12] as this does differentiate between a protein derived amine group and amine groups in small molecules.
Mass spectrometry analysis on the other hand is more sensitive and more specific and has allowed the measurement of melamine content, as well as detecting the presence of carcinogenic dyes (in Chilli powders) [11,12], plastic originating chemicals that interfere with the endocrine system and protein degradation products in mishandled meat [13].
Proteomics has been used to study the nutritional value of cereals, (the major food group which sustains the human race), comparing different species with the idea of customised nutrition [14]. This is akin to customised drug treatment after genotyping patients for specific cancer-related mutations, but instead of aiming to treat a disease after it has occurred, it aims to supply the best source of energy for the specific requirements of an individual.
A new branch of proteomics known as metaproteomics is also adding an insight into varied populations of microbes from soil samples to mucosal samples [15,16]. It describes the populations of microbes qualitatively by utilising multi-species databases. This information allows the description of a micro-niche population and could be applied to either crop and soil health, or human pathologies involving our micro-flora.
Human biomarker research is a less than trivial pursuit, not least due to large variations in abundance of key proteins in body fluids. In animals however biomarker discovery has proven more successful. McLean and colleagues discovered the tried and tested early warning system utilised in the feline kingdom to warn off unfriendly foes by proteomic analysis of cat urine. Cats, both domestic and of the large wild variety, excrete small molecules and proteins when spray marking (usually on up-right surfaces, to delineate territories or to leave dating invitations to the opposite sex). McLean and colleagues described the presence of the protein Cauxin in domestic cats, Sumatran tiger, Asiatic lion, clouded leopard and jaguar. Cauxin shares sequence homology with mammalian carboxylesterases, and is speculated in this investigation to cleave a pre-cursor of the feline urine protein felinine. Felinine is itself broken up into signalling pheromones for mating. Interestingly, they also found that Cauxin was present in the urine of cats that don’t produce felinine- indicating it may be a general esterase/peptidase used for processing the signals transmitted by all cats producing Cauxin [15].
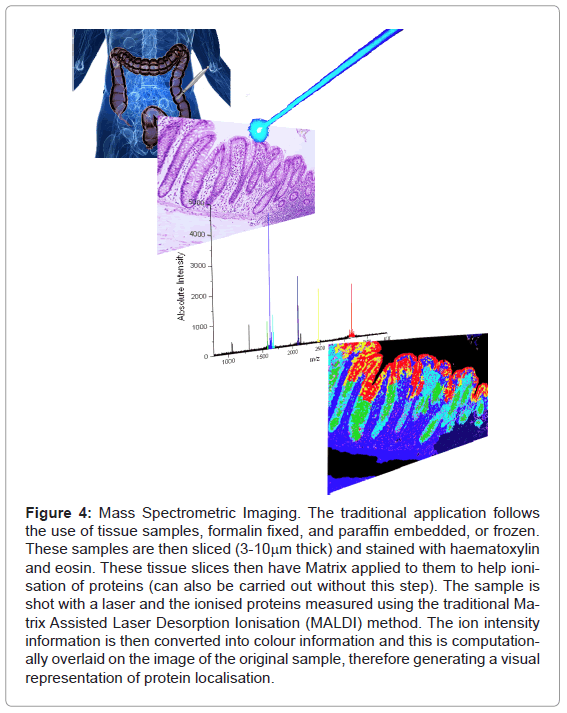
From mapping on a macro scale, proteomics and mass spectrometry has been used on the micro-scale as well. Traditional imaging with mass spectrometry allowed the ionisation of surface proteins on sections of tissue with a laser (Figure 4).
Figure 4: Mass Spectrometric Imaging. The traditional application follows the use of tissue samples, formalin fixed, and paraffin embedded, or frozen. These samples are then sliced (3-10mm thick) and stained with haematoxylin and eosin. These tissue slices then have Matrix applied to them to help ionisation of proteins (can also be carried out without this step). The sample is shot with a laser and the ionised proteins measured using the traditional Matrix Assisted Laser Desorption Ionisation (MALDI) method. The ion intensity information is then converted into colour information and this is computationally overlaid on the image of the original sample, therefore generating a visual representation of protein localisation.
From this technique individual proteins can be identified and a relative quantitation can be obtained. More generally however, it is used to globally measure the presence or absence of highly abundant proteins in each section of a target tissue/structure. Description of defined structures in particular tissue has been possible e.g. brain tissue sections or tumour margins. This is the typical use for this branch of proteomics and has spatial resolution in the sub 1μm range (TOF – SIMS -Time Of Flight Secondary Ion Mass Spectrometry [16]) to typically 50-70 μm (MALDI-MSI (Matrix Assisted Laser Desorption Ionisation- Mass Spectrometry Imaging)and SIMS-MSI (Secondary Ion Mass Spectrometry- Mass Spectrometry Imaging [17]). Novel medical uses of this technique are now allowing the diffusion and metabolism (or the movement and breakdown) of drugs in tissues to be characterised by their effectiveness to get to required targets and to assess the specificity of a drug for its designed target. The added benefits of proteomics in drug screening are the numerous secondary effects of the drug that can be characterised at the same time as looking at effects on specific target proteins. The multi-factorial nature of identifying and measuring the abundance of hundreds to thousands of proteins in an experiment gives proteomics an advantage over previously used assays and technologies. The potential for characterising drug effects over time, in the cell is also now being carried out on a more regular basis, by applying drugs to cells in culture, making drug testing more descriptive and ultimately safer at an earlier stage in the drug trial process.
While MALDI-TOF MS imaging is generating images for medicine, TOF-SIMS is being applied to the micrometer dissection of priceless works of art. Rembrandt’s Portrait of Nicolaes Van Bambeeck, 1641 has been analysed using TOF-SIMS imaging to determine the method of paint layer application (as you can analyse layer by layer with a Bismuth liquid metal ion gun as opposed to the laser source utilised in medical and standard MALDI-TOF) [16]. By analysing the peptide/ protein, polysaccharide, lipid and small molecule (metal ions etc.) components of the spectra obtained from samples of his paintings it has been possible to establish that some dye materials must have come from waste textile manufacture (which was not previously known), and additionally chemical reaction products, which could only have resulted from the combination of white lead and oils found in a binding medium used by oil paint artists.
The precise, scientific analysis of artwork may be taking away some of the emotive quality of an artwork (in the eyes of the artistof course the ‘how’ and the ‘why’ are the beauty in the eyes of the scientist!), but similar analysis has brought mass spectrometry and proteomic techniques into the courtroom. The initial purpose of using TOF-SIMS to analyse the quality of ink jet printing on paper, and comparisons of which inks print the best images [18,19] had the unexpected but fortunate by-product of the ability to characterise the depth of penetration into the paper. This meant there was a characterisation of ink components in the x, y and now z dimensions. From this commercially driven research came the ability to take a piece of evidence from a document alteration court case, and scientifically determine what had been written in which time sequence (http://pubs. acs.org/cen/science/89/8923sci2.html), due to the ionisation of only the uppermost layer of ink, in any situation where one section of writing may have been written over or altered by a second ink. Fingerprints over the top of the ink for analysis do confound the results however, but do lead to the next forensic application of Mass Spectrometric proteomic analysis.
Fingerprint collection and analysis has long been common practice for the police force in providing evidence of individuals involved in a crime scene, due to the unique patterns, which comprise a fingerprint. The excretions of skin are also characteristic of an individual and defining the proteome of a person’s fingerprint is now possible given their levels of sebaceous, apocrine, eccrine secreted proteins and/ or small molecules, which may be distinctive when compared to another individual. What has also proven possible with TOF-SIMS is the visualisation of elicit substances on finger print evidence [20]. Visualised the presence of amphetamine, methamphetamine and ecstasy on finger print samples taken from a number of different surfaces, describing a discerning pattern of occurrence which can not only place a person at a crime scene but implicate handling of illicit substances – which without this kind of evidence would otherwise be presented as circumstantial evidence.
MALDI-TOF and TOF-SIMS imaging does however have its limitations. It relies heavily on the ionise-ability of surface molecules, and abundance - so low abundance proteins are not detected with these kinds of analysis, and the more complex the sample is the more difficult the peak assignment may be. Often these MS imaging techniques are accompanied by the highly sensitive and descriptive Electron Spray Ionisation tandem Mass Spectrometry techniques (ESI-MS/MS), which along with a capability to analyse highly complex samples (such as cell lysates consisting of hundreds or thousands of types of proteins) give peptide sequence information - adding confidence to any protein ID made, and allowing for de-novo sequencing (or characterising novel proteins/peptides not curated in available databases). Specialised protein preparation can allow organellar separation, and therefore spatial proteome characterisation. This doesn't have the 1 micrometre resolution in the 2D image sense, as described above, but what the technique lacks in this aspect it gains in a clever use of metabolic labelling. Boisvert et al. [21] have utilised SILAC (Stable Isotope Labelling of Amino acids in Cell culture [22]) labelling in combination with cellular fractionation to investigate the intracellular distribution of proteins. Boisvert and colleagues were able to describe the relative location of over 2000 proteins between the nucleus, nucleolus and cytoplasm. They also found after DNA damage was induced they could detect changes in the steady state distribution of proteins in the cell in response to the induced stress. Using Pulse SILAC methodologies they were also able to characterise the turnover of over 8,000 proteins [23]
Inter-organism proteomic imaging has also recently been investigated. A particularly exciting application of this kind of imaging has been applied to bacterial competition, between Bacillus subtilis and Staphylococcus aureus [24]. The interaction of these two species is relevant to humans for several reasons. B. subtilis are very common bacteria found on nearly all bodily surfaces, and are actually added to foods and cosmetics due to the benefits these bacteria have in maintaining a healthy “body flora”. They have this effect because they protect their own environment by out-competing other bacteria in the immediate area. The process by which B. subtilis affects this control on other potentially pathogenic microbes is crucially reliant on spatial and therefore directional interactions. The controlling effects of B. subtilis on S. aureus, particularly methicillin-resistant S. aureus (the notoriously hard to treat hospital and nursing home acquired infection cause, MRSA) had not been spatially characterised until Gonzalez et al. utilised the MS imaging techniques which have only recently allowed this kind of analysis. They found that there was a distinct directionality of the antibiotics and metabolites by growing the competing microbes in a T conformation (S. aureus along the vertical of the T and B. subtilis along the horizontal), and then using a raster grid approach characterised the protein abundance at the interface between the 2 bacterial colonies. They found that in and around the S. aureus lawn there were detectable levels of the membrane disrupting delta toxin (of S. aureus origin), but in the area of S. aureus growth inhibition caused by B. subtilis, they found non-ribosomal circular peptides Surfactin and Plipastatin, which have been characterised as antibiotics. This was the first directional characterisation of inter-microbial warfare, and will enable targeted drug discovery, utilising nature’s well established and efficacious defence systems.
Circular peptides are generated by plants, animals and bacteria and are often completely independent of DNA and ribosomes. Many important and commonly used pharmaceuticals are actually cyclic peptides, including cyclosporine (an immune-suppressant), actinomycin D (an antibiotic) and Daptomycin (a lipopeptide antibiotic which targets MRSA). These molecules circumvent the “central dogma” of biology – DNA is transcribed into RNA- which is translated into Protein. Cyclic peptides generated by cyanobacteria (Figure 5) were found to be the cause of toxicity in algal blooms which have been responsible for wildlife, human, and shell fish poisoning.
What is more interesting is the evolutionary relevance of cyanobacteria, possibly the first life forms on earth. There is evidence of cyanobacteria existing on earth up to 3.5 billion years ago and were likely the cause of the oxygenation of the atmosphere, consequently making the planet suitable for more complex life [25]. It is possibly an interesting coincidence that "primordial origin of life" supporting molecule generation experiments, generate amines and amino acids [26,27].
Circular peptides, therefore could be interpreted as potentially the first complex structures generated by conjugation of those amino acids. If you consider further the function of most of these cyclic peptides – toxins and antibiotics, or defence mechanisms and/or quorum sensing, these are interaction and population based communications, albeit not terribly amicable- but communication none-the-less. Aside from the interesting evolutionary implications the MS analysis of cyclic peptides is technically challenging because of the non-linearity of the cyclic peptide- therefore, traditional amino acid sequencing of spectra is not always possible. Also they often contain modified amino acids, not found in commonly occurring proteins, such as methyl –alanine found in the cyclic peptide Seglitide (Figure 6) [28].
Proteomics not only has new implications for biological and forensic applications, it is pushing the boundaries of computation, database generation and curation, and statistics. The application of high throughput techniques, in proteomics has resulted in experiments that generate large volumes of data. For example, currently one sample, run on a mass spectrometer for ~100 minutes, can produce raw data files up to 2.5GB. This rate of data generation quickly builds up to a very large scale and given the on-going improvements in instrumentation this trend is only likely to increase in future. Many funding bodies currently supporting proteomics research have policies that necessitate the storage of these data for a minimum of 10 years. Simple storage of individual data files is not the only challenge, however, especially as the cost of large scale storage devices continues to fall. The real challenge is to make very large volumes of data easily accessible for continued analysis. Movements supporting this idea include the PRIDE data base (http://www.ebi.ac.uk/pride/). This is a public database to which scientists carrying out mass spectrometry based proteomics can submit meta data and mass spectra to, and contains 196,469,637 Spectra (identified + unidentified) [30]. The main aim of this public database was to cumulatively verify spectra and the resulting peptide and protein identifications, giving more statistical strength due to numbers of submitted spectra.
Mass spectrometry data alone do not provide answers to key questions in the applications described previously, it must be interpreted and analysed. The process of converting the MS data to information and then understanding, i.e. converting it to applicable knowledge, requires sophisticated methods of data processing and analytics. These methods must take into consideration the multi-faceted, complex nature of the global study of proteomes, including such parameters as how much of a protein is in a cell (intensity and abundance), where it is located in a cell (spatial localisation), who it talks to (interactions), and what chemical modifications it may gain or lose. Hence, when storing these data, traditional relational databases can be considered, however more recently multi-dimensional databases have been shown to play a role in providing a structure for proteomics data that allows for fast dimensional analytics to take place.
Boulon and colleagues presented a data analysis and storage platform for immuno-precipitation experiments [29]. These points to an emerging trend in proteomics, i.e. the use of large databases combined with sophisticated analysis software. The Boulon paper describes the first application of business intelligence strategies in proteomics to aid in the analysis of immuno-precipitation experiments. An immuno-precipitation experiment is the targeted isolation of a protein of interest, with the assumption that biologically interacting proteins will co-purify with the protein. This is the case, but often many non-specific proteins also co-purify in almost all experiments [30]. Using statistics related to the frequency of detection of proteins in multiple immuno-precipitation experiments helps to predict which protein identifications may be false. The Boulon et al. [29] paper describes a strategy of computationally combining experiments to give a frequency of detection for proteins in many immuno-precipitation experiments. Using this resource, helps to evaluate whether a protein identification in a new experiment might be a non-specific protein, or a genuine interaction partner (http://proteinfrequencylibrary.com). This application of business intelligence strategies provides proof of concept for combining multiple experiments in a structured data store to aid statistical evaluation of future experiments. Furthermore, using computational analysis techniques in this way allows us to combine large volumes of data to better address the complexity of proteomics, and use our historical knowledge in analysis of new experiments.
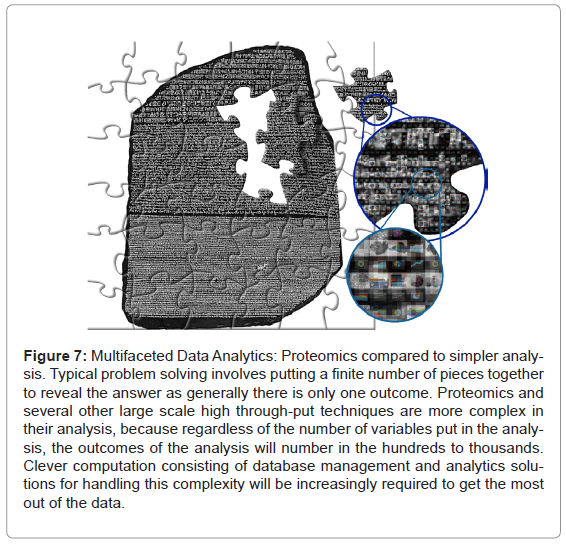
The specific application described above is an indicator of how potentially moving proteomics into the data analytics arena can help improve the use of high throughput multi-dimensional sciences like genomics and proteomics. Looking to the future of proteomics, with manual annotation no longer feasible, we envisage that this kind of data handling and interrogation has to become commonplace. Furthermore, collecting data systematically can help to build a baseline of historical knowledge that can be accessed repeatedly and utilised to find patterns and trends only apparent across multiple experiments over time (Figure 7).
Figure 7: Multifaceted Data Analytics: Proteomics compared to simpler analysis. Typical problem solving involves putting a finite number of pieces together to reveal the answer as generally there is only one outcome. Proteomics and several other large scale high through-put techniques are more complex in their analysis, because regardless of the number of variables put in the analysis, the outcomes of the analysis will number in the hundreds to thousands. Clever computation consisting of database management and analytics solutions for handling this complexity will be increasingly required to get the most out of the data.
Proteomics as a developing, and by nature high-tech, multidisciplinary science is having a wide- spread effect on humanity. Thanks to many complementary developments in methodology, cutting edge physical chemistry and engineering combined with the power of modern computing, proteomics offers huge future potential for answering major questions in biology and for providing new clinical advances leading to improved diagnoses and the safer treatment of human disease. The application of proteomics in these new fields (forensics, drug discovery, evolutionary research and food safety) is an indication of the growing relevance of this kind of analysis.
Acknowledgements
AIL is a Wellcome Trust Principal Research Fellow.
References
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, et al. (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnology & genetic engineering reviews 13: 19-50.
- Hu S, Loo JA, Wong DT (2006) Human body fluid proteome analysis. PROTEOMICS 6: 6326-6353
- Reymond MA, Schlegel W (2007) Proteomics in cancer. Adv Clin Chem 44: 103-142.
- Venugopal A, Pandey A, Chaerkady R (2009) Application of mass spectrometrybased proteomics for biomarker discovery in neurological disorders. Ann Indian Acad Neurol 12: 3-11.
- Gobezie R, Millett PJ, Sarracino DS, Evans C, Thornhill TS (2006) Proteomics: Applications to the Study of Rheumatoid Arthritis and Osteoarthritis. J Am Acad Orthop Surg 14: 325-332.
- Pan C, Kumar C, Bohl S, Klingmueller U, Mann M (2009) Comparative Proteomic Phenotyping of Cell Lines and Primary Cells to Assess Preservation of Cell Type-specific Functions. Molecular & Cellular Proteomics 8: 443-450.
- Branigan T (2008) Chinese figures show fivefold rise in babies sick from contaminated milk. The Guardian UK.
- Roberts AEKaMG (1905) A Method for the Direct Determination of Organic Nitrogen by the Kjeldahl Process. Public Health Pap Rep 31: 109-122.
- Shea F, Watts CE (1939) Dumas method for organic nitrogen Industrial & Engineering Chemistry Analytical Edition 11: 333-334.
- Moore JC, DeVries JW, Lipp M, Griffiths JC, Abernethy DR (2010) Total Protein Methods and Their Potential Utility to Reduce the Risk of Food Protein Adulteration. Comprehensive Reviews in Food Science and Food Safety 9: 330-357.
- Aiello DLD, Gionfriddo E, Naccarato A, Napoli A, Romano E, et al. (2001) Review: multistage mass spectrometry in quality, safety and origin of foods. European Journal of Mass Spectrometry 17: 1-31.
- Breidbach A, Bouten K, Kroeger K, Ulberth F (2010) Capabilities of laboratories to determine melamine in food - results of an international proficiency test. Analytical and Bioanalytical Chemistry 396: 503-510.
- Carbonaro M Proteomics: present and future in food quality evaluation. Trends in Food Science & Technology 15: 209-216.
- Kussmann M, Panchaud A, Affolter M (2010) Proteomics in Nutrition: Status Quo and Outlook for Biomarkers and Bioactives. Journal of Proteome Research 9: 4876-4887.
- McLean L, Hurst J, Gaskell C, Lewis J, Beynon R (2007) Characterization of Cauxin in the Urine of Domestic and Big Cats. Journal of Chemical Ecology 33: 1997-2009.
- Sanyova J, Cersoy S, Richardin P, Lapre´vote O, Walter P, et al. (2011) Unexpected Materials in a Rembrandt Painting Characterized by High Spatial Resolution Cluster-TOF-SIMS Imaging. Analytical chemistry 83: 753-760.
- Solon E, Schweitzer A, Stoeckli M, Prideaux B (2010) Autoradiography, MALDIMS, and SIMS-MS Imaging in Pharmaceutical Discovery and Development. The AAPS Journal 12: 11-26.
- Sodhi RNS, Sun L, Sain M, Farnood R (2008) Analysis of Ink/Coating Penetration on Paper Surfaces by Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS) in Conjunction with Principal Component Analysis (PCA). The Journal of Adhesion 84: 277-292.
- Filenkova A, Acosta E, Brodersen PM, Sodhi RNS, Farnood R (2011) Distribution of inkjet ink components via ToF-SIMS imaging. Surface and Interface Analysis 43: 576-581.
- Szynkowska MI, Czerski K, Rogowski J, Paryjczak T, Parczewski A (2010) Detection of exogenous contaminants of fingerprints using ToF-SIMS. Surface and Interface Analysis 42: 393-397.
- Boisvert F-M, Lam YW, Lamont D, Lamond AI (2010) A Quantitative Proteomics Analysis of Subcellular Proteome Localization and Changes Induced by DNA Damage. Molecular & Cellular Proteomics 9: 457-470.
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, et al. (2002) Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Molecular & Cellular Proteomics 1: 376-386.
- Boisvert F-M, Ahmad Y, Gierlinski M, Charrière F, Lamond D, et al. (2011) A quantitative spatial proteomics analysis of proteome turnover in human cells. Molecular & Cellular Proteomics.
- Gonzalez D, Haste N, Hollands A, Fleming T, Hamby M, et al. (2011) Microbial Competition Between Bacillus subtilis and Staphylococcus aureus Monitored by Imaging Mass Spectrometry. Microbiology.
- Olson J (2006) Photosynthesis in the Archean Era. Photosynthesis Research 88: 109-117.
- Parker ET, Cleaves HJ, Dworkin JP, Glavin DP, Callahan M, et al. (2011) Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proceedings of the National Academy of Sciences 108: 5526-5531.
- Miller S (1953) A production of amino acids under possible primitive earth conditions. Science 117: 528-529.
- Liu WT, Ng J, Meluzzi D, Bandeira N, Gutierrez M, et al. (2009) Interpretation of tandem mass spectra obtained from cyclic nonribosomal peptides. Analytical chemistry 81: 4200-4209.
- Boulon S, Ahmad Y, Trinkle-Mulcahy L, Verheggen C, Cobley A, et al. (2010) Establishment of a protein frequency library and its application in the reliable identification of specific protein interaction partners. Mol Cell Proteomics 9: 861-879.
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, et al. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. The Journal of Cell Biology 183: 223-239.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 17118
- [From(publication date):
August-2013 - Apr 10, 2025] - Breakdown by view type
- HTML page views : 12325
- PDF downloads : 4793