Editorial Open Access
Prokaryotic Biodiversity in Marine versus Terrestrial Ecosystems: Methylobacteria and Research Ethics
U. Kutschera* and S. SchauerInstitute of Biology, University of Kassel, D-34132 Kassel, Germany
- *Corresponding Author:
- Ulrich Kutschera
PhD, Institute of Biology, University of Kassel
Heinrich-Plett-Str.40, D-34132 Kassel, Germany
E-mail: kut@uni-kassel.de
Received date August 23, 2012; Accepted date August 24, 2012; Published date August 29, 2012
Citation: Kutschera U, Schauer S (2012) Prokaryotic Biodiversity in Marine versus Terrestrial Ecosystems: Methylobacteria and Research Ethics. J Marine Sci Res Dev 2:e113. doi:10.4172/2155-9910.1000e113
Copyright: © 2012 Kutschera U, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
Prokaryotic microbes (bacteria) are the most numerous organisms on Earth. In marine environments, cyanobacteria of the genus Prochlorococcus and heterotrophic prokaryotes of the family Pelagibacteraceae are of importance, and in terrestrial ecosystems, plant-associated methylobacteria are very abundant. Here, we summarize the discovery and description of a microbe isolated from the common cord moss, Methylobacterium funariae, as described by Schauer and Kutschera (2011). Based on samples of M. Funariae which was provided to the colleagues as a gift, our isolate was described without our knowledge and permission for a second time, under another name. We discuss this wasteful and duplicative publication of data generated twice from the same bacterial isolate with respect to the ethics for the conduct of research in the biological sciences.
Despite their minute size, bacteria are the most numerous and diverse forms of life in the world. In marine ecosystems, phototrophic cyanobacteria of the genus Prochlorococcus and heterotrophic microbes of the family Pelagibacteraceae, also known as the ‘SAR11 bacterial clade’, are among the most abundant prokaryotes [1]. However, despite their proliferation throughout the world’s oceans, the taxonomy of these important bacteria is still unclear. In terrestrial ecosystems, members of the alpha-proteobacterial genus Methylobacterium are abundant epiphytic microbes that have been isolated from the leaf surfaces of bryophytes, mosses, ferns, and seed plants. Methylobacteria are capable of growing on a variety of one-carbon compounds, such as methanol, which is emitted from the stomatal pores of the leaves. About 40 bacterial species (i.e., defined strains) of these pink-pigmented, Gramnegative prokaryotes have been described so far [2]. In this personal account, we describe a case study of bacterial systematics that sheds light on current problems in publication ethics [3].
In 2003, we isolated methylobacteria from the upper surface of the thallus of the liverwort Marchantia polymorpha. The “primitive” land plants were collected from Bergpark Wilhelmshöhe (Kassel, Germany). Later, we described one of our lab strains, labelled ‘JT1’, as novel species (Methylobacterium marchantiae) [4].
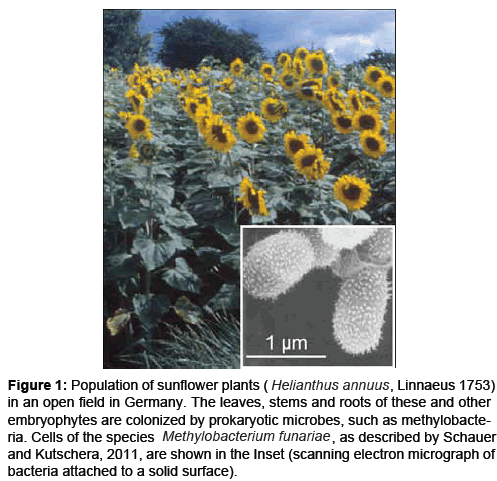
One year before (May 2002), we had isolated another strain (F3.2) from the phylloids of the common cord moss Funaria hygrometrica, collected in Botanical Garden of the University of Kassel, where sunflower plants were raised (Figure 1), and cultivated these microbes in the Plant Physiology laboratories of the Institute of Biology. This isolate was described informally as Methylobacterium funariae in the PhD thesis of the second author [5], and later, supplemented by additional data, as a new species in a peer-reviewed journal [6].
Figure 1: Population of sunflower plants (Helianthus annuus , Linnaeus 1753) in an open field in Germany. The leaves, stems and roots of these and other embryophytes are colonized by prokaryotic microbes, such as methylobacteria. Cells of the species Methylobacterium funariae,as described by Schauer and Kutschera, 2011, are shown in the Inset (scanning electron micrograph of bacteria attached to a solid surface).
In his “Validation list no. 145”, Euzéby [7] proposed that our strain F3.2 should be assigned to the taxon Methylobacterium bullatum sp. nov [8], but added the following footnote: “Based on the strain history of the type strain of Methylobacterium bullatum, it appears that strain F3.2 has also been used as the type strain of ‘Methylobacterium funariae’ (Schauer and Kutschera, 2011)” [6]. In May 2012, we received a pertinent e-mail from a colleague (D. P. Kelly, University of Warwick, UK; 15th February, 2012): “I have a query about Methylobacterium funariae. While making a survey of the 16S rRNA gene accessions of all the published species, I found that strain F3.2 has been described twice with different names (M. funariae and M. bullatum) by separate groups at Kassel. Can you tell me how this happened, and which of the names is likely to be validated in the International Journal of Systematic and Evolutionary Microbiology (IJSEM)? I am wondering if the earlier report (April 2011) will have taxonomic precedence, or whether the report by Hoppe et al. [8] will be the validated name, as they deposited the type strain in the German Collection of Microorganisms and Cell Cultures (DSMZ), as required by the international nomenclatural rules”. Moreover, a recent report [9] misinterpreted the two-fold description of our isolate F3.2, and erroneously discussed “two” separate bacterial species. However, our unpublished DNA sequencebased phylogenetic trees document that M. funariae and M. bullatum are identical bacterial species. In this editorial, we elucidate the mysterious two-fold description of our isolate Methylobacterium F3.2 (Figure 1).
In our first research paper that was published in the year when we isolated lab strain Methylobacterium F3.2, we characterized the unique phenotype of our “Methylobacterium sp. that was isolated from the phylloid” [10]. Moreover, we quantified the ability of Methylobacterium sp. F3.2 to secrete cytokinines, using Funaria hygrometrica-protonemata-bioassay. In two subsequent reports [11,12], Methylobacterium sp. F3.2 was further characterized with respect to its capability to synthesize and secrete the phytohormone auxin. Based on these extensive studies, “Methylobacterium sp. isolated from phylloids of F. hygrometrica” [10] was described as M. funariae [5,6].
In our article which was published ten years ago [10], we depicted scanning electron micrographs that document the rugged outer surface of Methylobacterium sp. F3.2 (see Figure.1, Inset). To further characterize the fimbriae-like structures on the outer wall layer of this isolate, we provided samples of Methylobacterium sp. F3.2 to the microbiology lab of the Institute of Biology, University of Kassel (Principal Investigator: F. Schmidt). However, without our knowledge and permission, our colleagues used our un-described microbial taxon to work on a second species description. They were informed that our isolate had already been named M. funariae by Schauer (2009) [5], and that a corresponding species description was in preparation. It should be noted that the first author of this second paper [8] proposed to share data, publish a joint, and give a valid species description in the IJSEM. We agreed to the proposal of T. Hoppe, but the PI of the microbiology lab (F. Schmidt) declined our offer (the second “author” on the Hoppe et al. [8] is a technician without academic qualification).
As a result, two species descriptions were published [6,8], and although our paper appeared in print first, and Schauer (2009) [5] had already described this taxon two years earlier, the name “M. bullatum” has been accepted by IJSEM [7]. We are aware of the fact that according to the Bacterial Code 1990 Revision, the “date of effective publication does not determine priority” (Rules 23b and 27).
In a recent editorial published in the IJSEM, Kämpfer [13] described a case of falsification of ‘Certificates of Deposit’ and reminded the readers that the IJSEM is a member of the Committee on Publication Ethics. We feel that the “M. Bullatum case” described here is clearly a violation of the ethical standards in biology, since, according to the “Codes and policies for research ethics”, scientists should avoid duplicate publication and not deceive colleagues [3]. We hope that such a wasteful and two-fold description of the same bacterial isolate, provided by the discoverers of this taxon as a gift to colleagues, will not happen again.
Finally, we want to point out that methylobacteria are among the most abundant heterotrophic microbes of the above-ground phytosphere (stems, leaves) in terrestrial ecosystems. Moreover, they also inhabit the rhizosphere of all land plants investigated so far [14]. Hence, these microbes may be viewed as ecological counterparts to the marine Pelagibacteriaceae, which are likewise members of the alphaproteobacteria [1,14]. However, more experimental work is required to further corroborate our conclusion that the marine SAR11 bacterial clade [1] and land plant-associated methylobacteria [14] are infact the dominant heterotrophic prokaryotic microbes in their respective ecosystems.
References
- Brown MV, Lauro FM, Demaere MZ, Muir L, Wilkins D, et al. (2012) Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8: 595.
- Green PN (2006) Methylobacterium In: Bergey's Manual of Systematic Bacteriology 567-571. Edited by Brenner DJ, Krieg NR, Garrity GM, Staley JT, Boone DR, de Vos P, Goodfellow M, Rainey RA, Schleifer KH: Springer-Verlag, New York.
- Shamoo AE, Resnik DB (2009) Responsible Conduct of Research (2ndedn). National Science Foundation: Oxford University Press, New York.
- Schauer S, Kämpfer P, Wellner S, Spröer C, Kutschera U (2011) Methylobacterium marchantiae sp. nov., a pink-pigmented, facultatively methylotrophic bacterium isolated from the thallus of a liverwort. Int J Syst Evol Microbiol 61: 870-876.
- Schauer S (2009) Biofilmbildung bei Pflanzen-assoziierten Bakterien der Gattung Methylobacterium und molekulargenetisch-physiolgische Charakterisierung eines neuen Marchantia-Isolats (Mtb. sp. JT1). Inaugural-Dissertation, Universität Kassel, Kassel.
- Schauer S, Kutschera U (2011) A novel growth-promoting microbe, Methylobacterium funariae sp. nov., isolated from the leaf surface of a common moss. Plant Signal Behav 6: 510-515.
- Euzéby J (2012) Validation List no. 145. List of new names and new combinations previously effectively but not validly, published. Int J Syst Evol Microbiol 62: 1017-1019.
- Hoppe T, Peters K, Schmidt F (2011) Methylobacterium bullatum sp. nov., a methylotrophic bacterium isolated from Funaria hygrometrica. Syst Appl Microbiol 34: 482-486.
- Tani A, Sahin N, Matsuyama Y, Enomoto T, Nishimura N, et al. (2012) High-throughput identification and screening of novel Methylobacterium species using whole-cell MALDI-TOF/MS analysis. PloS One 7: e40784.
- Hornschuh M, Grotha R, Kutschera U (2002) Epiphytic bacteria associated with the bryophyte Funaria hygrometrica: Effects of Methylobacterium strains on protonema development. Plant Biol 4: 682-687.
- Hornschuh M, Grotha R, Kutschera U (2006) Moss-associated methylobacteria as phytosymbionts: an experimental study. Naturwissenschaften 93: 480-486.
- Kutschera U (2007) Plant-associated methylobacteria as co-evolved phytosymbionts: a hypothesis. Plant Signal Behav 2: 74-78.
- Kämpfer P (2010) Certificates of deposit - a key element of the Bacteriological Code and an indispensable prerequisite for comparative taxonomic research. Report of a case of falsification and a reply to the letter to the editor by Tindall (2008). Int J Syst Evol Microbiol 60: 475-477.
- Schauer S, Kutschera U (2008) Methylotrophic bacteria on the surfaces of field-grown sunflower plants: a biogeographic perspective. Theory Biosci 127: 23-29.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 14562
- [From(publication date):
December-2012 - Oct 21, 2025] - Breakdown by view type
- HTML page views : 9862
- PDF downloads : 4700