Research Article Open Access
PiPE, a Phytophthora-associated PAMPS from P. infestans, Binds to a Ca2+-Dependent Protein Kinase (CDPK) in Potato for the Induction of Hypersensitive Reaction
Naotaka Furuichi1,2,*, Kazutoshi Yokokawa2, Hisakazu Okamura2 and Masatoshi Ohta21Department of Agriculture, Niigata University, Niigata, 950-2181, Japan
2Graduate School of Science and Technology, and Niigata University, Niigata, 950-2181, Japan
- *Corresponding Author:
- Naotaka Furuichi
Department of Agriculture, Niigata University
Niigata, 950-2181, Japan
Tel: +81-25-262-7520
Fax: +81-25-262-7520
E-mail: nfuru@agr.niigata-u.ac.jp
Received date: August 21, 2013; Accepted date: January 07, 2014; Published date: January 09, 2014
Citation: Furuichi N, Yokokawa K, Okamura H, Ohta M (2014) PiPE, a Phytophthora-associated PAMPS from P. infestans, Binds to a Ca2+-Dependent Protein Kinase (CDPK) in Potato for the Induction of Hypersensitive Reaction. J Clin Exp Pathol 4:156. doi:10.4172/2161-0681.1000156
Copyright: © 2014 Furuichi N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Abstract
We report that PiPE, a Phytophthora-associated PAMPS (Pathogen Associated Molecular Patterns), induced generation of active oxygen species and hypersensitive cell death (HR) by treatment of potato tuber tissues, and that the PiPE gene from a species of Oomycete, Phytophthora infestans Mont (de Bary), was cloned. Interaction of a His-tagged PiPE from P. infestans with a His-tagged Ca2+-Dependent Protein Kinase (RiCDPK2) from potato cv. Rishiri (R1-gene) was investigated by using enzyme-linked immunosorbent assay with mouse monoclonal anti-PiPE Antibodies (Abs). We found that the PiPE and a Mycelical Homogenate (MH) from P. infestans can interact with His- RiCDPK2 in vitro. PiPE showed binding interaction with the His-fusion proteins from three other domains of RiCDPK2, indicating the existence of binding sites for PiPE of P. infestans on RiCDPK2. We suggest that after binding with the PiPE, RiCDPK2 may trigger signals that lead to the occurrence of HR in potato.
Keywords
Phytophthora infestans; Hypersensitive cell death; CDPK2; Enzyme-linked immunosorbent assay; PiPE elicitor
Introduction
The interaction between plants and an incompatible pathogen leads often to rapid and localized cell death in the context of a hypersensitive response. Hypersensitive cell death (HR), a defense mechanism, is characterized by rapid cell death at the site of infection, which restricts further growth of the infecting fungus. The mechanisms of these molecular events are presumed to be as follows: (1) initial recognition of the PAMPS(pathogen associated molecular patters) and the suppressor of the pathogen by host plasma membrane in the infection process [1-3]; (2) increase in Ca2+ influx [4-6], decrease in pH, and kinase activation in the cells [4,7-11]; and (3) induction of biochemical and physiological defense in the host cells [1,10,12,13].
A PAMP of an Oomycete, Phytophthora Infestans (PiPE), and the Mycerial Wall Component (MWC) elicited Hypersensitive Reaction (HR) [13]. The reaction induced by the HWC from P. infestans in potato tuber was very similar to the defense response of that induced by infection with P. infestans [8,13,14].
In our long standing attempt to explore the antigenic potential of P. infestans-derived surface structures to elicit cultivar-non-specific defence response in potato, we have previously identified various proteinaceous cell wall fractions to contain active PAMPs [1,13]. PiPE, a mycerial wall derived PAMP, derived from a surface-existing glycoproteins of 38 kD P. infestans Fructose-1,6-Bisphosphate Aldolase (FBA) was shown to serve as a recognition PAMP for the activation of HR (hypersensitive cell death) and the generation of Active Oxygen Species (AOS) [1,13].
PiPE was almost invariably found to be associated with FBA from six Phytophthora species. The evolutionary conservation of PiPE together with its indispensability for both PAMP elicitor and enzyme activity of the intact protein reveals it as a genus-specific pathogen recognition similar to that described for many species from various kingdoms [4,7,15,16] Receptor binding of PiPE evokes a PAMS-specific cytoplasmic streaming, and the brownian movement in the cytosol, production of AOS as well as post translational activation of CDPK kinases, all of which are important elements for the transmission of the PiPE PAMP signal [1,13-15,17]. Now, we have isolated a gryco-protein PAMP from P. infestans, PiPE, which is found also in the wide species of Phytophthora spp., i.e. P. cinnamomi, P. megasperma, P. nicotianae and P. cryptogea.
Using this PiPE as PAMP of potato late blight enabled us to study the molecular cascades of converging signaling for HR or for inhibition of HR in potato system.
In contrast, the suppressor inhibited the interaction of HR in potato and P. infestans [8,18,19]. The signals from the elicitor and/ or suppressor of HR have been investigated. However, we previously proposed that plasma membrane Ca2+-Dependent Protein Kinase (CDPK) plays an important role in the signal pathway of the PAMPs from P. infestans in potato cells [1,2,13].
In the present study, using Enzyme-Linked Immunosorbent Assay (ELISA) with monoclonal anti-PiPE antibodies, we investigated whether the PiPE, Mycelical Wall Component (MWC) and the Mycelical Homogenate (MH) from P. infestans can interact with the His-tagged protein of Ca2+-Dependent Protein Kinase (RiCDPK2) from potato cv. Rishiri (R1-gene).
The nucleotide sequence of the PiPE from Phytophthora infestans and the RiCDPK2 have been deposited in the DDBJ/EMBLE/Genbank data bases under the accession numbers AB051573 (PiPE), and AB051809 (RiCDPK2).
Materials and Methods
Plant and CDPK genes
The cells from potato cv. Rishiri, an interspecific hybrid between Solanum tuberosum L. and S. demissum L., cultured in MS liquid medium [20] , were used for the cDNA isolation of RiCDPK2 [14]. The MWC and MH were extracted from P. infestans (Mont.) de Bary isolate DN101 (race 0) grown in rye-seed medium [19] according to the method described [21] and by Furuichi and Suzuki [18].
The His-tagged PiPE was prepared from P. infestans as reported [1,2,8,13]. The mouse monoclonal anti-PiPE antibodies-2A11 (PiPEAbs) used in this study described by Furuichi et al. 2013 [13] recognize a PiPE present in MWC from P. infestans.
The preparation of His-RiCDPK2
For the preparation of fusion proteins from RiCDPK2, the full length domain including kinase (K) I to X with junction- (J) and Ca2+-bindingdomain (C) called KJC, the kinase domain VIb to X with junction- and Ca2+-binding-domain (KVIbJC), the kinase-I to X with junction- (KJ) and junction- with Ca2+-binding-domain (JC) were cloned to pCRexpression vectors. The fusion proteins were expressed in BL21pLysS cells of E. coli and purified as described by Xpress TM System protein purification protocol (Invitrogen, The Netherlands). His-tagged proteins from different domains of RiCDPK2, obtained from various steps of preparation were separated by electrophoresis on a SDS-polyacrylamide (12%) gel and visualized with coomassie brilliant blue [14]. Description of K-J-Ca2+, KVIb-J-Ca2+, K-J and J-Ca2+ is described previously, corresponding to the CDPK gene structure stated in Table 1.
| Kinase I | Kinase III | |
|---|---|---|
| SdCDPK1 | LKEFFSIGKKLGQGQFGTTFKCVEKATGKEYACKSIAKRKLLTDDDVEDV | RREVQIMHHLAGHPHVISIKGAYEDAVAVHVVMEFCAGGELFDRIIQRGH |
| (Solanum demmisum) | ||
| NtCDPK2 | LKEFFSIGKKLGQGQFGTTFKCVEKATGKEYACKSIAKRKLLTDDDVEDV | RREVQIMHHLAGHPNVISIKGAYEDAVAVHVVMEYCAGGELFDRIIQRGH |
| (Nicotiana_tabacum ) | ||
| ZmCPK11 | LKDKYSLGRRLGQGQFGTTHLCVERATGKELACKSILKRKLGSDDDVEDV | RREIQIMHHLAGHPSVVGIRGAYEDAVAVHLVMELCGGGELFDRIVRRGH |
| (Zea_mays) | ||
| OsCDPK | LKDLYTIGKKLGQGQFGTTYLCVEKATGREFACKSIAKRKLLTQEDVEDV | RREIQIMHHLAGHANVVSIVGAYEDAVAVQLVMELCAGGELFDRIIQRGH |
| (Oryza_sativa ) | ||
| SdCDPK1 | YTERKAAELTRTIVGVVEACHSLGVMHRDLKPENFLFVDQKEDSLLKAID | FGLSIFFKPGDRFTDVVGSPYYVAPEVLRKRYGPEADVWSAGVILYILLS |
| NtCDPK2 | YTERKAAELTRTIVGVLETCHSLGVMHRDLKPENFLFVNQKEDSLLKTID | FGLSIFFKPGDRFNDVVGSPYYVAPEVLKKRYGPEADVWSAGVIVYILLS |
| ZmCPK11 | YTERKAAELARVIVGVVEACHSMGVMHRDLKPENFLFADHSEEAALKTID | FGLSIFFRPGQIFTDVVGSPYYVAPEVLKKRYGPEADVWSAGVIIYILLC |
| OsCDPK | YSEKAAAQLARVIVGVIEACHSLGVMHRDLKPENFLFIHQKEDSPLKAID | FGLSIFFKPGETFTDVVGSPYYVAPEVLMKHYGREVDVWSAGVIIYILLS |
| SdCDPK1 | GVPPFWAENEQGIFEQVLHGDLDFKSDPWPSISEDAKDLMRRMLVRDPRR | RLTAHEVLCHPWVQVDGVAPDKPLDSAVLSRMKQFSAMNKLKKMALRVIA |
| NtCDPK2 | GVPPFWAENEQGIFEEVLHGDLDFSSDPWPSISEDAKDLVRRMLVRDPKK | RLTAHEVLCHRWVQVDGVAPDKPLDSAVLSRMKQFSAMNKLKKMALRVIA |
| ZmCPK11 | GVPPFWAENEQGIFEEVLHGRLDFESEPWPSISDGAKDLVRRMLVRDPRK | RLTAHEVLRHPWVQVGGVAPDRPLDSAVLSRMKQFSAMNKLKKMALRVIA |
| OsCDPK | GVPPFWDESEQGIFEQVLKGDLDFSSEPWPNISESAKDLVRKMLIRDPKK | RLTAHEALCHPWVCVDGVAPDKPLDSAVLSRLKQFSAMNKLKKMALRVIA |
| Tether region | ||
| SdCDPK1 | ESLSEEEIAGLKEMFKMIDTDNSGQITFEELKEGLKRFGSNLKETEIYDL | MQAADVDNSGTIDYGEFIAATLHMNKIERQDHLFAAFCYFDKDGSGYITA |
| NtCDPK2 | ESLSEEEIAGLKEMFRMIDTDNSGQITFEELKVGLKRVGANLKESEIYDL | MQAADVDNSGTIDYGEFIAATLHFNKIEREDHLFAAFSYFDKDGSGYITA |
| (ZmCPK11) | ENLSEDEIAGLREMFKMIDADNSGQITFEELKVGLEKVGANLQESEIYAL | MQAADVDNNGTIDYGEFIAATLHLNKVEREDHLFAAFQYFDKDGSGYITA |
| OsCDPK | ESLSEEEIAGLKEMFKMLDTDNSGHITLEELKTGLQRVGANLMDSEIDAL | MEAADIDNSGTIDYGEFIAATLHINKVEKEDKLFAAFSYFDKDGSGYITQ |
| SdCDPK1 | DELQQACEEFGIGDVRMEEMIREADQDNDGRIDYNEFVAMMQK | |
| NtCDPK2 | DELQQACEEFGIGDVHLEDMIRDADQDNDGRIDYNEFVAMMQK | |
| ZmCDPK11 | DELQVACEEFGLGDVQLEDLIGEVDQDNDGRIDYNEFVAMMQK | |
| OsCDPK2 | DELQKACEEFGIGDTRIEDIIGDIDQDNDGRIDYNEFVEMMQ |
Table 1: Cluster analysis of CDPKs of various plant species.
The gene structure and the cDNA of CDPK2
The longest open reading frame (ORF) of RiCDPK2 cDNA, from kinase domain I to X with junction- and Ca2+ binding-domain, gener-ally called the EF hand (KJC, 1488 bp), and the three other domaincombinations of RiCDPK2 were amplified through PCR. The other three combinations of different domains included the kinase -domain VIb to X with junction- and EF hand (KVIbJC, 1050 bp), the kinase domain I to X with junction- (KJ, 966 bp) and the junction-domain with EF hand (JC, 615 bp). The oligonucleotide primers (5’to 3’) of the four samples designed to amplify the desired sequences for cloning are: KJC forward: 5´(ATGGAACCAAAACCAGCAACTG), KJC reverse: 5´(TAAGATTTCTTCACTCTGTACGAG); KVIbJC forward: 5´(ATGGGTGTTATGCATAGAGAT), KVIbJC reverse: (as of KJC); KJ forward: (as of KJC), K-J reverse: 5´(AATCACACGTAAAGCCATTTTCTT); JC forward: 5´ (ATGGCTCCTGATAAACCTCTT) and JC reverse: (as of KJC).
The PCR was carried out according to the standard conditions at 95°C for 5 min then 35 cycles 95°C for 30 s, 56°C for 30 s and 72°C for 1.75 min followed by a final extension at 72°C for 10 min. The PCR products were cloned to the pCR-expression vector with TOP 10F´ E. coli cells as host for characterization of the construct. The cloning was confirmed by using PCR with T7 forward and V5 C-terminus reverse primers.
Enzyme-Linked Immunosorbent Assay (ELISA)
Enzyme-Linked Immunosorbent Assay (ELISA) was performed according to the method described by Furuichi et al. (1990) [13,18] and McLaughlin et al. (1989) [22]. In the first step, the microtiter plate (Falcon) wells were coated with 250 ng of purified fusion proteins from KJC (59 kDa), KVIb-JC (43 kDa), KJ (40 kDa), and JC (27 kDa) domain combinations of RiCDPK2, and diluted with coating buffer (15 mM Na2CO3, 35 mM NaHCO3, 0.02% NaN3, pH 9.6). The plate was incubated overnight at 4°C. The antigen was removed and the wells were blocked using 100 ul of phosphate-buffered saline (PBS) (0.12 M NaCl, 2.7 mM KCl, 3.2 mM Na2HPO4.12H2O) with 3% bovine serum albumin (BSA), and the plate was incubated at 37°C under cover for 1 hr. The wells were washed 5 times with PBST (PBS containing 0.5% polyvinylpyrrolidone and 0.1% Tween-20). The respective wells were coated with 250 ng per 50 ul of the antigens, which contained fusion, PiPE, MWC, and MH from P. infestans diluted with the coating buffer. The plate was incubated at 37°C for 1 hr. The wells were washed 5 times with PBST and treated with 50 É l of PiPE-Abs diluted at 1:2000 in PBS (with 1% BSA and 0.05% NaN3). The plate was incubated at 37°C for 1 hr. After five washes with PBST, 50 ul of alkaline phosphataseconjugated rabbit anti-mouse antibodies (DAKO) diluted at 1:1000 in PBS (with 1% BSA and 0.05% NaN3) were applied to the wells, followed by incubation at 37°C for 1 hr. In the case of the control treatments, the wells were respectively coated with PiPE, MWC, and MH, and with coating buffer only in the next step and then detected with PiPEAbs. Finally, the wells were washed with PBS, and 50 ul of the enzyme substrate p-nitrophenyl phosphate tablet (5 mg, Sigma) diluted in 5 ml of substrate buffer (1 M Diethanolamine, 1 mM MgCl2, 0.04% NaN3, pH 9.8) was applied to each well. Absorbance was read at 405 nm on a microplate reader (BioRad 550) after 1 hr.
Results and Discussion
The partial sequence alignment of the PiPE from P. infestans was shown in Figure 1 after [23]. It was shown that PiPE-gene has 674 bp. Figure 1 contained seven other FAB-genes from fungus (PiPE: peptide elicitor of P. infestans, Um: Ustilago maydis, Sc: Saccharomyces cerevisiae, Nc: Neurospora cassa, Pm: Pasteurella multocida, Dh: Debaryomyces hansenii, Cg: Candida glabrata, Hi: Haemophilus influenzae).
Figure 1: Partial sequence alignment of the PiPE from P. infestans. Electrophoresis showed PiPE-gene (674 bp) after [13], Alignment analysis of PiPE-gene from P. infestans. It was shown that seven other FAB-genes were reported from fungus. Partial nucleotide sequence of the mRNA encodes the PiPE protein from P. infestans and deduced amino acid sequence. The arrowhead indicates predicted glycosylation sites. The alignments were made with the program DNAsys. Residue letters are indicated under the sequences. (PiPE: Peptide elicitor of P. infestans, Um: Ustilago maydis, Sc: Saccharomyces cerevisiae, Nc: Neurospora crassa, Pm: Pasteurella multocida, Dh: Debaryomyces hansenii, Cg: Candida glabrata, Hi: Haemophilus influenzae).
The PiPE gene showed the identity, 100%, with the XM 00290, Phytophthora infestans T30-4 fructose bisphosphate aldolase gene.
His-tagged CDPK2, H-KJC, H-KJ and H-JC binds with PiPE
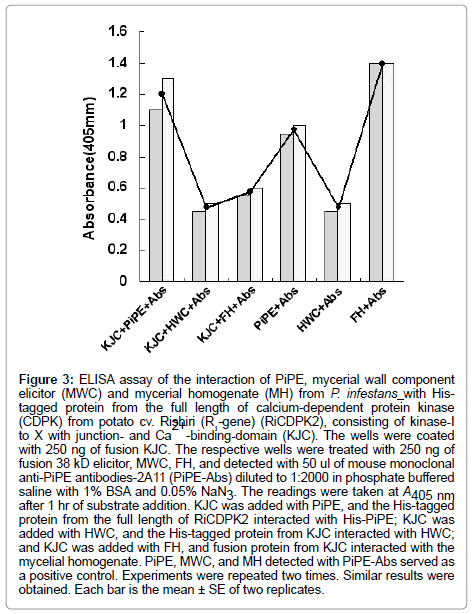
In the present experiment, the His-tagged proteins (250 ng) and His-tagged PiPE PAMP (250 ng) were assessed for interaction. In the first experiment, His-PiPE, MWC, and MH from P. infestans showed interaction with KJC (kinase-domain, Junction- and Ca2+ bindingdomains) of CDPK2 after 15 min of substrate addition at 23°C. Strong binding was detected in the cases of PiPE and KJC interaction (Ab, 1.178) after 1 h of incubation at 23°C (Figure 2). We found that the absorbance of the interaction of MWC (Ab, 0.48) and MH (Ab, 0.615) with KJC of CDPK2 was lower than PiPE with KJC (Ab, 1.178). It was observed that PiPE showed greater binding activity to KJC of CDPK2 than did MWC of MH of P. infestans, which was obvious from the control interactions of PiPE with PiPE-Abs (Ab, 0.983) and MWC with PiPE-Abs (Ab, 0.500). The high absorbance (Ab, 1.383) in the case of the MH with PiPE-Abs was assumed to be due to some other glycoprotein molecules attaching to the antigen in MH, corresponding to the PiPEAbs, or being detected by PiPE-Abs. We analyzed the interaction of PiPE with the other three subdomains of His-tagged proteins of RiCDPK2 (Figure 3). The PiPE showed positive interaction with the His-tagged proteins from KVIbJC, KJ, and JC. Strong signals were detected for PiPE against KVIbJC (Ab, 0.888), KJ (Ab, 0.932), and JC (Ab, 0.923), though these signals were lower than that of the interaction of PiPE with KJC (Ab, 1.178). The His-tagged-KJC (59 kDa) is larger than the three in the other CDPK domains (43, 40, and 27 kDa). Interestingly, it was shown that at the same concentrations of His-tagged proteins, the larger the size of the protein, the stronger its absorbance of interaction with PiPE. Positive interaction of PiPE with the His-tagged proteins from different domains of RiCDPK2 shows that there may be Kinase domain-I and -III of RiCDPK2 that act as binding sites for PiPE from P. infestans as previously reported d [8]. Similarly, the high absorbance of the interaction of PiPE with KJC as compared with that of the other three smaller His-tagged proteins from RiCDPK2 may be due to the presence of several kinase domains which could bind with the binding sites on full length RiCDPK2. We reported previously that the kinase domain I and III of RiCDPK2 effected by the Abs for each domains, and that they reduced their kinase activity by the binding of Abs [8]. The comparison of the alignment of CDPKs from various plant species were shown in Table 1. They were compared between Kinase domains (K), Junction domains (J), and EF-hands (EF), calcium binding domains (C) in Table 1.
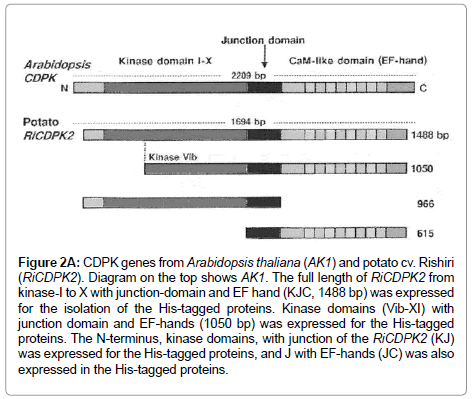
Figure 2A: CDPK genes from Arabidopsis thaliana (AK1) and potato cv. Rishiri (RiCDPK2). Diagram on the top shows AK1. The full length of RiCDPK2 from kinase-I to X with junction-domain and EF hand (KJC, 1488 bp) was expressed for the isolation of the His-tagged proteins. Kinase domains (Vib-XI) with junction domain and EF-hands (1050 bp) was expressed for the His-tagged proteins. The N-terminus, kinase domains, with junction of the RiCDPK2 (KJ) was expressed for the His-tagged proteins, and J with EF-hands (JC) was also expressed in the His-tagged proteins.
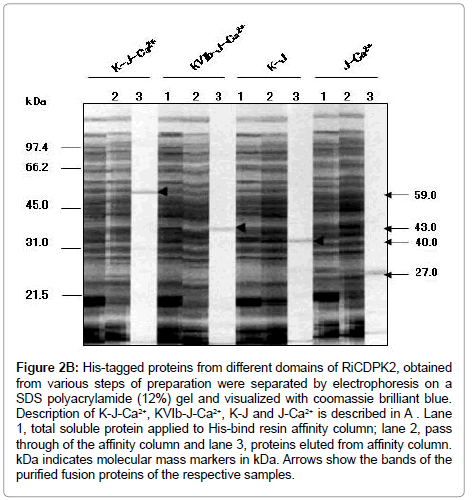
Figure 2B: His-tagged proteins from different domains of RiCDPK2, obtained from various steps of preparation were separated by electrophoresis on a SDS polyacrylamide (12%) gel and visualized with coomassie brilliant blue. Description of K-J-Ca2+, KVIb-J-Ca2+, K-J and J-Ca2+ is described in A . Lane 1, total soluble protein applied to His-bind resin affinity column; lane 2, pass through of the affinity column and lane 3, proteins eluted from affinity column. kDa indicates molecular mass markers in kDa. Arrows show the bands of the purified fusion proteins of the respective samples.
Figure 3: ELISA assay of the interaction of PiPE, mycerial wall component elicitor (MWC) and mycerial homogenate (MH) from P. infestans with Histagged protein from the full length of calcium-dependent protein kinase (CDPK) from potato cv. Rishiri (R1-gene) (RiCDPK2), consisting of kinase-I to X with junction- and Ca 2+ -binding-domain (KJC). The wells were coated with 250 ng of fusion KJC. The respective wells were treated with 250 ng of fusion 38 kD elicitor, MWC, FH, and detected with 50 ul of mouse monoclonal anti-PiPE antibodies-2A11 (PiPE-Abs) diluted to 1:2000 in phosphate buffered saline with 1% BSA and 0.05% NaN3. The readings were taken at A405 nm after 1 hr of substrate addition. KJC was added with PiPE, and the His-tagged protein from the full length of RiCDPK2 interacted with His-PiPE; KJC was added with HWC, and the His-tagged protein from KJC interacted with HWC; and KJC was added with FH, and fusion protein from KJC interacted with the mycelial homogenate. PiPE, MWC, and MH detected with PiPE-Abs served as a positive control. Experiments were repeated two times. Similar results were obtained. Each bar is the mean ± SE of two replicates.
ELISA assay by using PiPE, MWC and MH, the pathogen homogenates
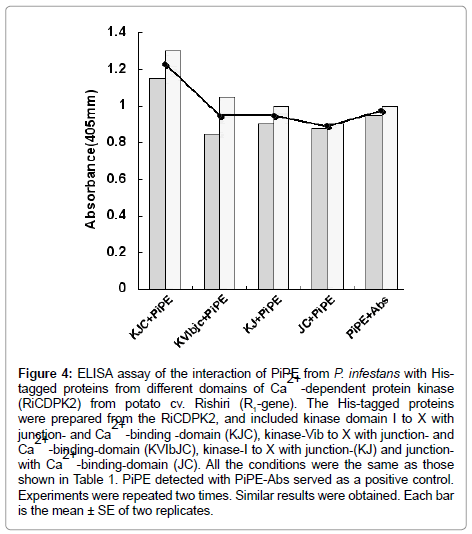
The present results show that PiPE, MWC, and MH can interact with the His-RiCDPK2 in vitro with different intensity than that shown in the ELISA assay (Figure 4). Moreover, we observed positive interaction of PiPE with His-tagged proteins from different domains of RiCDPK2. It was assumed that RiCDPK2 might contain multiple binding sites for PiPE from P. infestans. We suggest that the interaction of a PAMP with RiCDPK2 may initiate the signaling that leads to the occurrence of interaction between HR triggered potato and P. infestans interaction. We proposed the CDPK2 signaling in cytosol in infected cells can regulate the occurrence of hypersensitive cell death in potato cell.
Figure 4: ELISA assay of the interaction of PiPE from P. infestans with Histagged proteins from different domains of Ca2+ -dependent protein kinase (RiCDPK2) from potato cv. Rishiri (R1-gene). The His-tagged proteins were prepared from the RiCDPK2, and included kinase domain I to X with junction- and Ca2+ -binding -domain (KJC), kinase-Vib to X with junction- and Ca2+ -binding-domain (KVIbJC), kinase-I to X with junction-(KJ) and junctionwith Ca2+ -binding-domain (JC). All the conditions were the same as those shown in Table 1. PiPE detected with PiPE-Abs served as a positive control. Experiments were repeated two times. Similar results were obtained. Each bar is the mean ± SE of two replicates.
It was reported that ectopic expression of a heterologous CDPK (AK-6H) in tomato protoplasts enhanced plasma membrane-associated NADPH oxidase activity [24]. We examined the effect of CDPK antibodies, recognizing kinase domain-III, to Active Oxygen Species (AOS) generation in potato membrane fractions. It was observed that the treatment of CDPK antibodies to the membrane fraction of potato, which was expressed His-rboh1 protein, inhibited approximately 50% of AOS generation. This means that CDPK signaling in potato cells play an important role in the NADPH oxidase activation in potato plasma membrane [1,2,8,13].
Our present studies were consistent with the evidence that CDPK in potato was located upstream of NADPH oxidase in the host plasma membrane in the signal pathway that leads to the induction of AOS generation. We reported that CDPK1 and CDPK2 was localized in the membrane fraction of potato, and that Ca2+ stimulated the membrane kinase activity in potato [8,14]. These lines of evidence suggest that the activity of NADPH oxidase is regulated by the CDPK signaling after the PiPE binding to the kinase on potato plasma membrane [13]. This hypothesis is now underway by using a single photon-detecting crosscorrelation microscopy system with CDPK-1 and -2 in a single potato suspension cell.
Acknowledgements
We thank Drs. Anderson A (USU) for useful advice and A. Shirata for encouragement throughout this study. We also thank for Kato M and Sawahata R for preparation of the gene constructs, and Hasegawa Y for preparation of figures. NF was supported by the grants from the Ministry of Education, Science and Culture, by the grant from JSPS, Japan and supported by the grant from the Intelligent Cosmos Science Foundation (Sendai).
References
- Furuichi N (2012) PiPE, effector, and suppressor stimulate the Ca2+-dependent protein kinase cascades in hypersensitive response in plant cells. Proceedings of 2nd International Congress of Microbiology.
- Furuichi N, Yokokawa K (2010) A nobel PAMPS (PiPE) from Phytophthora infestans induces active oxygen species and the hypersensitive reaction in potato. Proceedings of 1st Asian Conference on Plant-Microbe Symbiosis and Nitrogen Fixation.
- Danna CH, Millet YA, Koller T, Han SW, Bent AF, et al. (2011) The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc Natl Acad Sci U S A 108: 9286-9291.
- Lee H, Chah OK, Sheen J (2011) Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473: 376-379.
- Kiba A (1995) Specific inhibition of cell wall bound ATPases by fungal suppressor from Mycosphaerella pinodes. Plant Cell Physiol 36: 809-817.
- Ebel J, Scheel D (1992) Elicitor recognition and signal transduction, in Genes involved in plant defense, Springer, Wien, Austria.
- Novacky A, Goodman R (1994) The Hypersensitive Reaction in Plants to Pathogen. APS press.
- Furuichi N, Yokokawa K (2008) PiP Elicitor and Suppressor from Phytophthora infestans Regulate Ca2+-Dependent protein kinase (CDPK) in the plasma membrane of potato. Japanese Journal of Plant Sciences, 2: 35-38.
- Greenberg JT (1996) Programmed cell death: a way of life for plants. Proc Natl Acad Sci U S A 93: 12094-12097.
- Qi Z, Verma R, Gehring C, Yamaguchi Y, Zhao Y, et al. (2010) Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc Natl Acad Sci U S A 107: 21193-21198.
- Xu H, Heath MC (1998) Role of calcium in signal transduction during the hypersensitive response caused by basidiospore-derived infection of the cowpea rust fungus Plant Cell 10: 585-598.
- Ricci P (1997) Induction of the hypersensitive response and systemic acquired resistance by fungal protein: The case of elicitins, Chapman and Hall, New York, USA.
- Furuichi N, Yukokokawa K, Okamura H, Ohta M (2013) A novel elicitor (PiPE) from Phytophthora infestans induces active oxygen species and the hypersensitive response in potato. Global Journal of Medical Research 8: 1-14.
- Furuichi N, Yokokawa K, Ichihara T (2008) Ca2+-dependent protein kinase in Tomato is Stimulated by Host-Selective Toxin from Alternaria solani. Plant Stress 2: 152-155
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, et al. (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418-422.
- Tomiyama K (1982) Hypersensitive cell death, in Plant Infection: The Physiological and Biochemical Basis, Japan Sci. Soc. Press, Tokyo/Springer-Verlag, Berlin, Germany.
- Peiter E (2011) The plant vacuole: emitter and receiver of calcium signals. Cell Calcium 50: 120-128.
- Furuichi N, Suzuki J (1990) Purification and properties of suppressor glucan isolated from Phytophthora infestans. Ann Phytopathol Soc Japan 56: 457-467.
- Doke N, Tomiyama K (1980) Effect of hyphal wall components from Phytophthora infestans on protoplasts of potato tuber tissues. Plant Pathol 16: 169-176.
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiological Plantrum 15: 473-497.
- Garas N.A, Doke N, Kuc J, (1979) Suppression of the hypersensitive reaction in potato tubers by mycelial components from Phytophthora infestans. Physiol Plant Pathol 15: 117-126.
- McLaughlin RJ, Chen TA, Wells JM (1989) Monoclonal antibodies against Erwinia amylovora: Characterization and evaluation of a mixture for detection by enzyme-linked immunosorbent assay. Phytopathology 79: 610-613.
- Feng XL (2011) Locally stabilized P-1-nonconforming quadrilateral and hexahedral finite element methods for the Stokes equations. Journal of Computational and Applied Mathematics 236: 714-727.
- Xing T (2001) Ectopic expression of an arabidopsis calmodulin-like domain protein kinase-enhanced NADPH oxidase activity and oxidative burst in tomato protoplasts. Molecular Plant Microbe Interaction 14: 1261-1264.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14262
- [From(publication date):
January-2014 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 9661
- PDF downloads : 4601