Research Article Open Access
PIK3CA/AKT1 Mutations in Breast Carcinoma: a Comprehensive Review of Experimental and Clinical Studies
Megan L. Troxell*Department of Pathology and Knight Cancer Institute, Oregon Health & Science University, USA
- *Corresponding Author:
- Megan L. Troxell
Department of Pathology, L471
3181 SW Sam Jackson Park Rd, Portland, OR 97239, USA
Tel: 503-418-1770
Fax: 503-494-8148
E-mail: troxellm@ohsu.edu
Received Date: November 21, 2011; Accepted Date: January 27, 2012; Published Date: January 30, 2012
Citation: Troxell ML (2012) PIK3CA/AKT1 Mutations in Breast Carcinoma: a Comprehensive Review of Experimental and Clinical Studies. J Clin Exp Pathol S1:002. doi: 10.4172/2161-0681.S1-002
Copyright: © 2012 Troxell ML. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Abstract
The phosphatidylinositol-3-kinase (PI3K) pathway is an important nexus for integration of extracellular and intracellular signals, and there are very frequent perturbations of this pathway in breast cancer, making it an attractive target for therapeutic manipulation. Hotspot mutations in PIK3CA or AKT1 are found in nearly 30% of breast cancers, especially estrogen receptor-positive and Her-2/neu-positive subgroups. This review will emphasize experimental models, clinical characterization and implications of PIK3CA/AKT1 mutations in breast cancer. In vitro studies have demonstrated that these mutations are kinase-activating and can confer cellular transforming properties in the correct context. Further, overexpression of PIK3CA H1047R in a variety of murine models results in mammary proliferation and carcinomas, and established carcinomas may become PIK3CA H1047R-independent. Data from human breast carcinomas regarding the clinicopathologic significance of PIK3CA/AKT1 mutations were contradictory at first, yet trends are beginning to emerge. PIK3CA mutation seems to impart a favorable prognosis in estrogen-receptor positive breast cancers, and mutations are seen early, in pre-invasive breast lesions. Although larger studies are needed, PIK3CA mutations may not confer selective advantage in the metastatic setting. Numerous pharmacologic compounds targeting the PI3K pathway are in development. The complexity of PI3K crosstalk with other signaling cascades, negative feedback regulation, and the myriad of other genotypic and phenotypic deviations in breast cancers argues for thorough molecular characterization of tumors in cancer trials.
Introduction
In 2004, Samuels and the Vogelstein-Velculescu group first characterized point mutations in the PIK3CA gene, encoding the 110α-catalytic subunit of phosphatidylinositol-3-kinase (PI3K) in a variety of human cancers [1]. Meta-analysis of the breast cancer literature reveals that PIK3CA mutations are present in 26% of invasive breast cancers, making this gene one of the most frequently mutated in breast cancer, with most mutations concentrated in ‘hotspots’ in the helical domain (exon 9) or the catalytic domain (exon 20), with a smaller number of mutations preferentially in exons 1, 4 and 7. (Figure 1, Table 1, and references therein). More recently, Carpten et al. [36] characterized mutations in the plekstrin homology domain of AKT1 (found in 3.8% of breast cancers), an important downstream kinase. These discoveries have stimulated a large body of exciting in vitro, in vivo, and translational studies investigating the scope and implications of PIK3CA and AKT1 mutations.
Phosphatidylinositol-3-kinase (PI3K) is a heterodimeric lipid kinase that plays a key signaling role in epithelial cells, as it directly transduces signals from receptor tyrosine kinases (such as EGFR, HER2, IGFR, PDGFR, integrins), and G-protein coupled receptors via Ras [2-8]. Besides mutations of PIK3CA and AKT1, which will be the focus of this review, other perturbations of this pathway in breast cancer include PIK3CA copy number gains (in 6-14% [9-11]), loss or mutation of the inhibitor phosphatase and tensin homolog deleted from chromosome 10 (PTEN, in 25% or more) [7,11,12], and rarely mutation of the p85 regulatory domain (PIK3R1), among others [13]. A number of recent reviews have covered the PI3K signaling cascade, as well as potential pharmacologic targeting of the pathway [2-8,14-20]; thus, this review will emphasize experimental cell line models, mouse mammary models, and studies of PIK3CA/AKT1 mutations in human breast carcinoma samples.
Phosphatidylinositol-3-kinase pathway
PI3K 110α (PIK3CA) catalyzes the addition of a phosphate to phosphatidylinositol (4, 5) bisphosphate (PIP2) at the 3-position to form the active second messenger phosphatidylinositol (3, 4, 5) triphosphate (PIP3). PIP3 recruits phosphoinositide-dependent protein kinase 1 (PDK1) and AKT to the cell membrane, where AKT is phosphorylated and activated by PDK1 and mTORC2 [4,5,7,8,15-17,19-21]. AKT signalling has far-reaching effects intracellularly, interacting with mTORC1 proliferative pathway, metabolic pathways, transcription factors (NFκB, mdm2, etc.), cyclin dependent kinases; inhibiting glycogen synthetase kinase-3, inactivating proapoptotic factors (BAD, procaspase 9, forkhead transcription factors); among other interactions [4,5,7,8,15-17,19-21]. Recent work has also revealed AKT independent PI3K signaling via the Bruton tyrosine kinase (BTK) pathway, the Tec non-receptor tyrosine kinase family, serum- and glucocorticoidregulated kinases (SGK), and small GTPase regulators that factor in to cell polarity and migration [8,22]. Further, there is crosstalk with a myriad of other intracellular pathways, including AMPK, MAP kinase, and estrogen receptor mediated pathways [4,5,7,8,15-17,19-21], and a complex network of feedback signalling, such that inhibition of AKT may induce the expression and phosphorylation of receptor tyrosine kinases [23]. While PTEN antagonizes signaling by dephosphorylating PIP3, careful dissection of signaling pathways reveals differential effects of PIK3CA mutation and PTEN loss [5]. The intricacies of the signaling pathway are detailed in several excellent manuscripts [4,5,7,8,15-17,19-21].
Activating PIK3CA mutations in vitro
Initial in vitro studies demonstrated that the helical domain hotspot mutations (exon 9 E542K, E545K), as well as the kinase domain hotspot mutation (exon 20 H1047R) resulted in increased lipid kinase activity of PI3K 110a, and demonstrated transforming properties in chick-embryo fibroblasts, chick embryos, and NIH 3T3 cells. [1,5,24-28] Further, some of the less common PIK3CA point-mutations also were shown to have activating properties, albeit weaker, in a chick embryo fibroblast model (strong phenotype: N345K, C420R, P539R, E545G,Q546K, Q546P, H1047L; intermediate phenotype: E545A, T1025S, M1043I, M1043V, H1047Y, weak phenotype: R38H, K111N) [5,28,29]. The same phenotype has been demonstrated in a number of mammalian mammary cell-culture lines, with the H1047R and E545 mutations imparting anchorage independent proliferation in soft agar, growth factor independent proliferation, and abnormal invasive morphogenesis in Matrigel cultures, with other mutations again showing a weaker phenotype along the same spectrum [5,28,30-32]. In contrast, Gustin et al. [33,34] expressed mutant PIK3CA alleles at physiologic levels in MCF10 or hTERT-HME1 breast cancer cell lines, and found that mutant expression alone was not sufficient for growth in soft agar or abnormal morphogenesis in matrigel, compatible with the notion that multiple ‘hits’ are required for carcinogenesis in vitro [33,34].
Study of double-mutant cell lines have provided further insight into the disparate effects of PIK3CA helical and kinase domain mutants. Mutations in exon 9 (helical domain), disrupt the constitutive inhibitory effect of the PI3K p85 subunit, but remain dependent upon interaction with RAS [5,32,35]. In contrast, exon 20 mutations (kinase domain), become independent of RAS, yet require binding to the p85 regulatory domain [5,19,32,35]. Interestingly, recent in vitro studies have also shown that PIK3CA mutant kinases, especially exon 9 helical domain mutants, may preferentially activate PDK1 and its substrate SGK3, rather than the canonical AKT pathway [22]. These studies suggest that the different PIK3CA hotspot mutations in human tumors could have different effects, likely also influenced by cell type, microenvironment, and interplay with other aberrant tumor signaling pathways.
Activating AKT1 mutations
Mutations in AKT family members in cancer remained elusive for many years, as none were found in studies of the kinase domain. However, in 2007 Carpten et al. [36] identified transforming mutations in the plekstrin-homology domain of AKT1, present in a number of cancers, including breast cancer. The plekstrin-homology domain is a key segment involved in AKT1 localization to the cell membrane, and interaction with PIP3. [36-38]The activating mutation E17K alters the lipid binding pocket, resulting in increased membrane localization, AKT1 activation, and transformation of NIH 3T3 or Rat1 cell lines [36]. Again, expression of mutant AKT1 at physiologic levels in breast epithelial cells lines did not prove sufficient for transformation [34]. The various isoforms of AKT have been linked to different cellular phenotypes, with AKT1 associated with cell survival and growth in an environmental dependent fashion, and AKT2 linked to invasiveness. There is concern that inhibition of AKT1 could result in compensatory AKT2 overactivation, thus promoting invasion and metastasis [2,8].
PIK3CA kinase domain mutations in murine models
Three mouse models of mammary specific overexpression of PIK3CA with the activating H1047R kinase domain mutation (exon 20) have recently been published; no exon 9 transgenic models have been published to date [39-41]. Adams et al. [39] utilized the ROSA26 knock-in system and MMTV-Cre to drive expression of either H1047R or wild type PIK3CA in the mouse mammary gland (in a 129/CD1 background, backcrossed to FVB). Two variant lineages were studied, with differences in strength of recombination/transgene expression levels, and specificity of restriction to the mammary gland. Female mice from both H1047R mutant lines developed mammary tumors starting at 5 months of age, and showed reduced survival, while animals overexpressing wild type PIK3CA remained healthy [39]. The mammary tumors in H1047 animals included adenomyoepitheliomas and adenosquamous carcinomas, which were shown to generally express estrogen receptor and to have elevated levels of phospho- AKT(S473), PTEN and phospho-c-Jun(S73) [39]. The authors also investigated interaction of PIK3CA H1047R mutation with p53 in double transgenic animals, and found and increased rate of tumor formation, and a different spectrum of tumor histology, predominantly poorly differentiated and spindle cell, as is fairly characteristic of p53 recombinant mouse models [39,42]. As a side note, some H1047R transgenic mice without Cre recombinase developed blood vessel lesions, attributed to spontaneous activation of H1047R expression in endothelial cells [39]. This ties in with observations of Graupera et al. [43] that PIK3α is involved in angiogenesis and endothelial migration [3,43].
Meyer et al. [40] generated PIK3CA H1047 and GFP expressing animals, with expression driven by either MMTV-Cre, or WAP-Cre recombination in mammary luminal epithelial cells, in a FVB/N background. Female mice from both lineages developed proliferative mammary gland abnormalities. Although 75% of the MMTV animals died at a young age of undetermined causes (attributed to MMTV promoter leakiness causing deleterious expression elsewhere), surviving animals developed mammary carcinomas at an average of 7 months of age [40]. In the WAP strain, parous mice developed tumors more rapidly than nulliparous females, as would be expected given enhanced WAP activity and thus recombination and transgene expression with pregnancy [40]. A delayed involutional phenotype with a dramatic decrease in mammary gland apoptosis was also demonstrated after pregnancy [40]. As noted in the Toronto mouse model, the mammary carcinomas in the transgenic animals were of a variety of histologic types, predominantly adenomyoepitheliomas and adenosquamous carcinoma, and showed increased phospho-AKT levels, with about 20% of the adenomyoepitheliomas expressing estrogen receptor.
An elegant transgenic mouse model with conditional overexpression of HA-tagged PIK3CA H1047R was created by Liu et al. [41,44] In this model, PIK3CA H1047R overexpression is driven by an inducible promoter, with expression of the reverse tetracycline transactivator linked to the MMTV promoter, resulting in transgene expression in the mammary gland in the presence of tetracycline (doxycycline) in a FVB background [41,44]. Prior studies with this system demonstrated titratable levels of reporter proteins, with somewhat increased expression during pregnancy/lactation, as MMTV is hormonally dependent, and slight heterogeneity of expression in older animals [44]. Like the other models, proliferative mammary gland lesions were seen in PIK3CA H1047R expressing animals, and carcinomas developed at an average of 7 months [41]. The tumors were predominantly of solid, mixed or squamous morphology, with acinar, glandular, and papillary histologies also seen. Tumors had abundant phospho-AKT (S473), and phospho-S6RP (S235, S236) by immunohistochemical analysis. Interesting, upon withdrawing doxycycline (turning PIK3CA H1047 expression off), about one-third of tumors completely regressed, but nearly two-third of the tumors partially regressed and then resumed growth [41]. These recrudescent tumors were essentially independent of the PIK3CA transgene, and several exhibited additional genetic alterations, including Met, cdkn2a or mvc, which also imparted resistance to a PIK3CA targeted compound, GDC-0941.[41] This model suggests that activating PIK3CA mutations may have an important role in tumorigenesis, but that additional ‘hits’ often occur, promoting PIK3CA oncogene independence and sustained tumor growth even upon inhibition of a key tumor-initiating pathway [41].
AKT1 E17K transgenic mouse mammary models have not yet been published. Prior studies with activated (myristated) AKT1 demonstrated accelerated tumorigenesis but reduced metastasis, and activated AKT2 increased metastasis in a background of activated ERBB2 or polyoma virus middle T antigen [45].
Common features of these three contemporary PIK3CA H1047R mouse models include robust development of mammary tumors upon mammary gland directed overexpression of the PIK3CA activating mutation H1047R. Further, each of the models showed a similar variety of murine tumor morphologies, and estrogen receptor expression reminiscent of human tumor data demonstrating PIK3CA mutations in a wide spectrum of breast carcinomas, as detailed below (Table 1; of note, estrogen expression is relatively rare in mouse mammary carcinoma as compared to human carcinoma) [46-48]. The authors have remarked on the prevalence of murine adenosquamous carcinomas in H1047R transgenic models, drawing analogy to the apparent high prevalence of PIK3CA mutations in human metaplastic breast carcinoma. However, it should be noted that mammary tumors in nongenetically engineered animals and some other genetic models have a predilection for squamous or adenomyoepitheliomatous morphology (FVB, BALB/c and C3H, and in association with chemical carcinogens; Wnt pathway transgenic) [42,48,49]. Other caveats include the supraphysiologic expression levels of the mutant transgene, and the presence of epitope tags in some of the models.
PIK3CA mutations in breast carcinomas
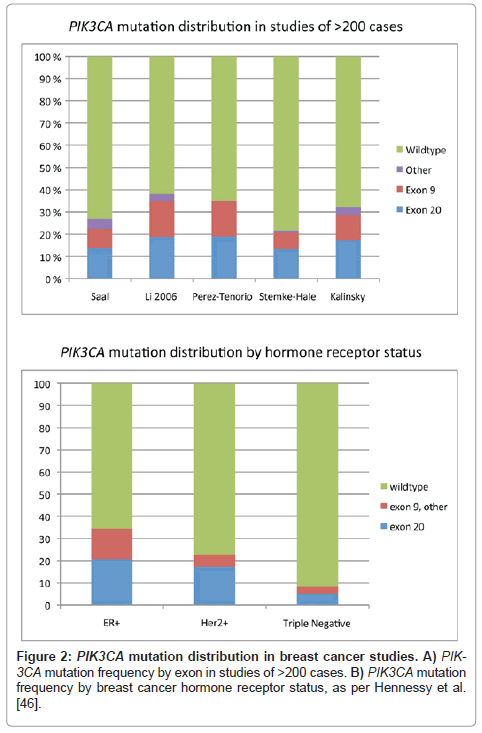
There is an extensive body of literature regarding the prevalence of PIK3CA mutations in resected human breast carcinoma specimens (Table 1, Figure 1-2). Meta-analysis indicates an overall PIK3CA mutation rate of 26% in breast carcinoma, including studies of different tissue substrates (frozen, formalin-fixed paraffin-embedded, methacarn-fixed paraffin embedded), tumor dissection methods, and different testing strategies interrogating various combinations of exons, with different sensitivities (Table 1 and references therein). Exon 20 kinase domain hotspot mutations comprise the majority of reported PIK3CA mutations in breast carcinoma (~57%), while exon 9 helical domain mutations comprise ~37%. Other exons and other protein domains such as the N-terminal adaptor binding domain, Ras binding domain, and the C2 putative membrane binding domain are more rarely altered (~6%, Figure 1) [5,50]. Rare individual cases have more than one PIK3CA mutation (Table 1) [51-58]. Many studies analyzed both normal breast and carcinomas from the same patients, and demonstrated PIK3CA mutations were restricted to tumor tissue (somatic), as mutations were not found in normal breast tissue [9,53,54,59-65].
| A. Unselected primary breast carcinomas | |||||||
|---|---|---|---|---|---|---|---|
| Author | PIK3CA Mutations | Exon | Method | mutation positive &clinical variables | |||
| # Mutations/ #Cases Tested | % | 9 | 20 | Other# | |||
| Samuels [1] | 1/12 | 8.3 | Sequence all exons | ||||
| Bachman [59] | 9/41 | 22.0 | 1 | 6 | 2 | Sequence exons 1, 9, 20 | |
| Campbell [60] | 28/70 | 40.0 | 15 | 9 | 4 | SSCP all exons & DHPLC exons 9, 20 | No association with ER, grade, LN |
| Saal [54] | 79/292^ | 27.0 | 26 | 40 | 13 | Sequence exons 1, 2, 4, 5, 7,9, 12, 13,18, 20 | ER+ PR+ Her-2/neu+ |
| Wu [9] | 19/92 | 20.6 | 13 | 6 | Sequence exon 1, 9, 20 (Frozen tissue) | ||
| Lee [51] | 26/78^ | 33.3 | 4 | 22 | SSCP exons 9, 20 | ||
| Levine 2005 [96] | 13/72 | 18.1 | 9 | 4 | Sequence exons 9, 20 (Frozen tissue) | ||
| Li [52] | 95/250^ | 38.0 | 40 | 47 | 8 | SSCP exon 7, 9, 20 (Frozen tissue) | ER+ PR+, size. Worse survival |
| Buttitta [61] | 46/180 | 25.6 | 23 | 23 | SSCP all exons & direct sequence exons 9, 20 (Frozen tissue) | Exon 9: lobular. No association with age, size, ER, PR, LN. | |
| Liang [53] | 33/80^ | 41.2 | 11 | 22 | Sequence exon 9, 20 | Age, stage. No association with ER. | |
| Maruyama [63] | 54/176 | 30.7 | 17 | 29 | 8 | Sequence exons 1, 2, 4, 7, 9, 18, 20 (Frozen tissue) | ER+, pAKT+ Favorable prognosis. |
| Perez-Tenorio [55] | 66/270^ | 24.4 | 30 | 36 | SSCP exon 9, 20 (Frozen tissue) | ER+ Her2-, small size | |
| Wood [95] | 5/11 | 45.4 | 3 | 2 | Full genome sequencing | ||
| Barbareschi [62] | 45/163 | 27.6 | 24 | 21 | SSCP exon 9, 20 (Frozen tissue) | Exon 20-improved overall and DF survival. Exon 9-worse overall and DF survival. | |
| Benvenuti [97] | 14/86 | 16.3 | 6 | 8 | Sequence exons 9, 20 | No association with age, ER, LN | |
| Lai [72] | 39/152 | 25.7 | 14 | 21 | 4 | Sequence exons 7, 9, partial 4, 20 | Exon 20 worse overall survival |
| Liedtke [98] | 23/140 | 16.4 | 12 | 11 | Sequence exon 1, 9, 20 (Frozen FNA) | Exon 9: LN-, ER+ | |
| Stemke-Hale [56] | 117/547^ | 21.4 | 40 | 73 | 7 | PCR-mass-spec (21 codons) (Frozen tissue) | ER+ Her2+, less common in basal-like. No association with outcome in tamoxifen treated ER+. |
| Leong [99] | 6/20 | 30 | 3 | 3 | Sequence exon 9, 20 | Two H1047R mutations, others non-hotspot | |
| Haverty [100] | 13/51 | 25.5 | 7 | 4 | 2 | Sequence all exons (frozen tissue) | Lack of mutation associated with chromosomal gain Her2 |
| Board [101] | 19/49 | 38.7 | 3 | 16 | ARMS-Scorpion (3 codons) (Frozen tissue) | ||
| Kalinsky [71] | 192/590 | 32.5 | 69 | 101 | 21 | PCR-mass-spec (17 codons) | ER+ Her2- LN- good prognosis; ex20:LN-, ex9:older age at dx |
| Miron [80] | 35/120 | 29.2 | 4 | 31 | Oncomap (12 codons) | ||
| Li [64] | 29/108 | 26.9 | Sequence exon 9, 20 (frozen tissue) | ER+ PR+ | |||
| Bozhanov [102] | 45/144^ | 31.3 | 18 | 28 | Sequence exon 9, 20 (Frozen tissue) | PR+, trend for better outcome | |
| Michelucci [68] | 68/176^ | 35.4 | 25 | 43 | Direct sequence exon 9, 20 (mostly frozen) | ER+ in Her2-; lobular subtype. No survival association | |
| Kadota [10] | 25/161 | 15.5 | |||||
| Jaiswal [50] | 14/62 | 22.5 | 2 | 10 | 2 | Sequence all coding exons | |
| Vorkas [65] | 62/174^ | 35.6 | 18 | 45 | High resolution melt analysis, exon 9, 20 | ||
| Dunlap [79] | 12/81 | 14.2 | 4 | 8 | DHPLC exon 7, 9, 20 | No association ER, Her2, LN | |
| Lopez-Knowles [11] | 12/168 | 7.1% | 4 | 8 | Sequence exon 9, 20 | No association with intrinsic cancer subtype | |
| Kan [50] | 29/183 | 15.8 | 27 | 2 | Mismatch Repair Detection (frozen tissue) | ||
| Boyault [103] | 22/120 | 18.3 | 10 | 12 | Sequence exon 9, 20 (frozen tissue) | ER+ | |
| Board [101] | 14/47 | 29.8 | 5 | 9 | ARMS (3 codons) | Control group for study of metastatic tumors | |
| Total | 1309/4966 | 26.3 | 460 | 725 | 73 | ||
| B. Studies of selected tumor phenotypes | |||||||
| Author | PIK3CA mutations | Exon | Methods | Tumor phenotype; PIK3CA mutation positive &clinical variables | |||
| # mutations/ positive &clinical variables cases tested |
% | 9 | 20 | Other | |||
| Berns [84] | 14/55 | 25 | 4 | 10 | Study of trastuzumab treated patients | ||
| Lerma[105] | 7/56 | 12 | 1 | 6 | Sequence exons 9, 20 | Study of Her2+, Triple Negative Associated with Her2+ and worse outcome |
|
| Hennessy [46] | 9/19 | 47 | 5 | 5 | PCR-mass-spec (~21 codons) (frozen tissue) | Study of metaplastic carcinoma | |
| DiNicolantonio [106] |
1/15 | 6.7 | 1 | Sequence exon 9, 20 | Study of advanced stage everolimus treated patients |
||
| Toi [27] | 5/29 | 17.2 | 3 | 2 | Sequence exons 9, 14-20 | Study of laptinib in Stage IIIb+ progressing on anthracyclines or taxanes |
|
| Michelucci [68] | 2/22 | 1 | 1 | Direct sequence exon 9, 20 (mostly frozen) | Hereditary breast cancers (both mutations in BRCA2, 0/10 BRCA1) |
||
| Ellis [73] | 127/398 | 32 | 46 | 85 | 1 | Sequence exon 9, 20 | Study of ER+ endocrine trial cases. Exon 20 improved DF survival |
| Loi [57] | 46/173^ | 26 | 14 | 32 | SSCP exon 9, 20 | Study of ER+/Her2- only. PIK3CA mutation gene signature good prognosis. |
|
| Esteva [12] | 12/55 | 21.8 | 5 | 7 | Sequencing exon 9, 20 | Study of Her2+ trastuzumab treated breast cancers |
|
| Da Silva [74] | 2/12 | 16.7 | 1 | 1 | PCR-mass-spec (13 codons) | Study of primary tumors with brain metastasis | |
| Dupont-Jensen [75] |
45/101 | 45 | 15 | 36 | SNaPshot PCR 3 codons | Study of primary tumors with subsequent metastasis |
|
| Cizkova^ [58] | 106/292 | 36.3 | 44 | 46 | Sequence exon 9, 20 (frozen tissue) Exons only provided for validation cohort (n- 249) |
Study of ER+ tumors. Characterizes PIK3CA mutation gene signature. |
|
| Razis [86] | 38/175 | 21.7 | Taqman SNP, 3 codons | Study of trastuzumab treated metastatic tumors. ER+, decreased time to progression |
|||
| Wang [87] | 7/57 | 12.2 | 2 | 5 | Sanger sequencing exon 9, 20 (2 non-hotspot mutations identified) |
Study of Her2+ trastuzumab, taxane and anthracycline treated. |
|
| Gonzalez-Angulo [76] |
19/47 | 40.4 | 1 | 18 | PCR-mass-spec (~21 codons) | Study of primary tumors with subsequent metastasis |
|
| Duprez [66] | 22/49 | 44.8 | 12 | 10 | PCR-mass-spec (13 codons) | Study of papillary carcinomas | |
| Duprez [66] | 14/49 | 28.5 | 1 | 13 | PCR-mass-spec (13 codons) | Study of low grade infiltrating ductal carcinomas |
|
| Board [101] | 10/45 | 23 | 7 | 3 | ARMS-Scorpion (3 codons) | Study of metastatic carcinomas | |
A. Unselected primary breast carcinomas
^identified tumors with 2 PIK3CA mutations: Lee-1 case, Li-7 cases, Liang-2 cases, Saal-2 cases, Perez-Tenorio-1 case, Stemke-Hale-4 cases, Loi-1 case, Cizkova-2
cases
COSMIC database (Catalog of Somatic Mutations in Cancer; http://www.sanger.ac.uk/genetics/CGP/cosmic/) lists 1360 mutations in 5402 tested breast carcinomas as of
10/30/11 (overall 25% mutation), including all of the above except: Li 2005, Maruyama 2007, Perez-Tenorio 2007, Haverty 2008, Hennessy 2009, Vorkas 2010, Dupont-
Jensen, Cizkova 2011, Wang 2011, Razis Gonzalez,-Angulo 2011, Duprez 2011; Board 2011.
NS-not significant; ND-not reported; DF-disease free; ER-Estrogen receptor; PR-Progesterone receptor; LN-Lymph nodes
DHPLC-denaturing high performance liquid chromatography; SSCP-Single strand conformation polymorphism; ARMS- Amplification Refractory Mutation System
Table 1: Literature review of PIK3CA mutations in invasive breast carcinomas.
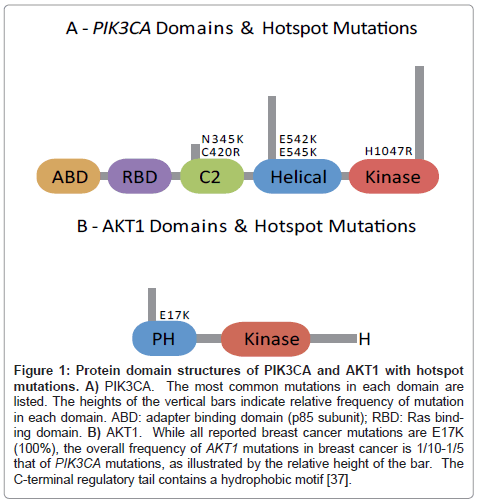
Figure 1: Protein domain structures of PIK3CA and AKT1 with hotspot mutations. A) PIK3CA. The most common mutations in each domain are listed. The heights of the vertical bars indicate relative frequency of mutation in each domain. ABD: adapter binding domain (p85 subunit); RBD: Ras binding domain. B) AKT1. While all reported breast cancer mutations are E17K (100%), the overall frequency of AKT1 mutations in breast cancer is 1/10-1/5 that of PIK3CA mutations, as illustrated by the relative height of the bar. The C-terminal regulatory tail contains a hydrophobic motif [37].
Early studies did not reveal correlation of PIK3CA mutation status with clinicopathologic parameters of carcinoma grade, stage, hormone receptor status, Her2 status, or outcome. However, with data from additional larger studies, several patterns have emerged. PIK3CA mutations are seen preferentially in estrogen receptor (ER)-positive and Her2/neu-positive tumors, and are less prevalent in ‘triplenegative' carcinomas (Table 1, Figure 2) [46]. However, Hennessy et al. [46] found a notable exception, in that their cohort of 19 squamous and spindle cell metaplastic carcinomas had a nearly 50% frequency of PIK3CA mutations. Lobular carcinomas also appear to have a relatively high rate of mutation [62,63]. Few of the other morphologic ‘special types of breast carcinoma’ have been studied in the literature; papillary and micropapillary carcinomas appear to have a mutation profile similar to ductal carcinomas, perhaps with enrichment of AKT1 mutations in micropapillary carcinomas (our preliminary data) [61,66]. In contrast, our group and others have found that mucinous carcinomas, which are often low-grade and ER+, have a paucity of PIK3CA mutations [52,60,61,63,67-69]. Further, we previous studied phyllodes tumors, and found no PIK3CA/AKT pathway mutations, while Vorkas reported one fibroadenoma with an H1047R mutation, suggesting a role in epithelial malignancy, and less of a role in fibroepithelial lesions [65,70].
PIK3CA mutation and breast cancer prognosis
Initial data was also contradictory regarding the association of PIK3CA mutations and breast cancer outcome, with studies of substantial numbers of patients demonstrating alternatively worse survival, or favorable prognosis of breast cancers harboring PIK3CA mutations.[52,63,71], Li et al. [52] reviewed a cohort of 250 breast carcinomas, and found a 35% frequency of PIK3CA mutations (exons 7, 9, 20). PIK3CA mutations were associated with worse survival (p=0.004); however, mutations were associated with larger tumor size (p=0.004) and ER-positivity (p=0.008) [52]. In multivariate analysis, PIK3CA mutation did not reach statistical significance in the overall group or in ER+ tumors, but was independently associated with worse survival in the Her2 negative group (p=0.016) [52]. In contrast, a Japanese study of 188 tumors found a 30.7% PIK3CA mutation frequency (exons 1, 2, 4, 7, 9, 13, 18, 20), and found mutations to be associated with favorable prognosis in uni- and multivariate analysis (p<0.05) [63]. Mutations were also significantly associated with ERpositivity and phospho-AKT positivity [63]. Similarly, a larger study from Memorial-Sloan Kettering demonstrated PIK3CA mutations in 32.5% of 590 breast cancers, and demonstrated that patients with PIK3CA mutated tumors have significantly improved overall (p=0.03) and cancer-specific survival (p=0.004), while mutations were associated with older age at diagnosis, hormone receptor positivity, Her2 negativity, lower grade and stage, and lymph node negativity [71]. However, the improvement in survival was independent of hormone receptor status [71]. This group also analyzed outcome based on type of PIK3CA mutation, and found that the survival benefit correlated with kinase domain (exon 20) mutation (p=0.005), but not helical domain (exon 9) mutation (p=0.54). They also noted kinase domain mutation to correlate with lymph node negativity (p=0.007), and helical domain mutation to correlate with older age at diagnosis (p=0.004) [71].
In addition to the Memorial Sloan Kettering group, other studies have separately analyzed the association of helical domain (exon 9) and kinase domain (exon 20) mutations with breast cancer outcome, which again yielded contradictory data.[62,72] An Italian study of 163 breast carcinomas found a PIK3CA mutation rate of 28%, with 53% of mutations in exon 9, and 47% exon 20 [62]; of note, most other studies have found a greater frequency of exon 20 mutations. Patients with exon 20 mutations had significantly better overall and disease free survival, as compared to patients without PIK3CA mutations [62]. Conversely, patients with exon 9 mutations had worse 5 year overall (p=0.018) and disease free (p=0.040) survival as compared to patients without PIK3CA mutations, and certainly as compared to patients with exon 20 mutations [62]. In multivariate analysis, the presence of exon 9 mutations maintained significance in terms of prognosis, and was the strongest predictor of disease free and overall survival (p=0.001 and p=0.0003 respectively), with nodal status and progesterone receptor expression also significant [62]. In contrast, a study from Taiwan found poor prognosis associated with exon 20 mutations [72]. This study documented a 26% PIK3CA mutation frequency in 152 breast cancer cases; of the mutations, 35% were in exon 9, 70% in exon 20, 12% in exons 4, 7 [72]. Exon 20 mutations were associated with worse overall survival at 5 years in univariate analysis (p=0.005), but not with disease free survival (p=0.5877) [72]. In multivariate analysis, the presence of PIK3CA exon 20 mutation was again a risk factor for significantly worse survival (p=0.0038) along with nodal status (p=0.0004) [72].
More recently, PIK3CA mutations have been analyzed in the context of somewhat more homogeneous subgroups of breast cancer, notably ER+ tumors [57,58,73]. These studies have generally revealed improved prognosis with PIK3CA mutation in this subset of breast cancer, a somewhat paradoxical finding considering that these are kinase activating mutations [57,58,73]. In a study of ER+ tumors from several different neoadjuvant endocrine therapy trials, Ellis et al. [73] demonstrated a 32% PIK3CA mutation frequency, and found presence of mutation to be a favorable prognostic factor for relapse free survival in univariate (p=0.02) and multivariate analysis, despite a weak negative correlation with clinical response to neoadjuvant endocrine therapy.
PIK3CA mutation gene signature
Using PIK3CA mutation status in addition to gene expression analysis of 173 ER+/Her2- breast carcinomas, Loi et al. [57] characterized a PIK3CA mutation “gene signature,” and then validated this signature in several other published datasets. In the initial dataset, 91% of PIK3CA mutations were exon 20 H1047R substitutions (overall 26% mutation rate). They found that PIK3CA mutation was indeed associated with a distinct gene expression profile, and in fact, some tumors showed this expression profile in absence of mutation, suggesting alternate mechanisms of pathway activation. Of note, the PIK3CA mutation-like gene signature appears to be distinct from the PTEN-loss expression signature. While PIK3CA mutated carcinomas were shown to have somewhat better outcome than wildtype carcinoma in the initial dataset, the PIK3CA-mutant like gene signature had still better prognostic significance [57]. The PIK3CA-gene signature was negatively correlated with signatures of proliferation, AKT activation, and PTEN loss, but positively correlated with ESR1 expression. Further protein data revealed decreased expression of mTOR, S6K and S6, with decreased phosphorylation of 4EBP1 and S6, in contrast to data from cell lines [57].
With a similar strategy, Cizkova et al. [58] characterized the transcriptome changes in PIK3CA mutated tumors. In this French cohort, 36% of cases had PIK3CA mutation, almost equally divided between exon 9 and exon 20 in the validation group of 249 tumors [58]. Differentially regulated genes amongst PIK3CA mutant and wildtype tumors were analyzed by cellular pathways, revealing impact on many signalling networks, including MAPK, Calcium, Jak-STAT, Wnt, apoptosis, with the Calcium pathway altered by downregulated genes, and Wnt pathway by upregulated genes [58]. This group further characterized a 29-gene set for discrimination of PIK3CA mutant and wild-type tumors, and verified it by RT-PCR in the 249 tumor cohort. Of note, 2 genes in this set were differentially regulated by exon 9 vs. exon 20 mutation. (TFAP2B-overexpressed in exon 20 mutated tumors and NRIP3-overexpressed in exon 9 mutated tumors). Interestingly, of the 29 discriminatory genes published by Cizkova, 6 are common to Loi’s 278 gene signature (Table 3) [57,58].
| Author | AKT1 E17K # Mutations/ Cases tested | % | Methods | Clinicopathologic correlation |
|---|---|---|---|---|
| Carpten 2007 [36] | 5/61 | 8.2% | Complete sequencing | 4/5 mutants ER+ |
| Bleeker 2008 [67] | 16/273 | 5.8% | Sequence hotspot | No clinical/hormonal data |
| Kim 2008 [106] | 4/78 | 5.1% | SSCP | No clinical/hormonal data DCIS: 0/15 |
| Stemke-Hale 2008 [56] | 6/418 | 1.4% | PCR-mass-spec | All mutants ER+ |
| Kalinsky 2009 [71] | 21/590 | 3.6% | PCR-mass-spec | Mostly ER+ |
| Kadota 2009 [10] | 11/161 | 6.8% | ||
| Lauring 2010 [34] | 3/100 | 3% | Sequence hotspot (frozen tissue) | All mutants ER+ |
| Dunlap 2010 [79] | 3/78 | 3.8% | Sequence hotspot | All mutants ER+ |
| Kan 2010 [50] | 4/183 | 2.2% | Mismatch Repair Detection (frozen tissue) | |
| Boyault 2011 [103] | 5/120 | 4.2% | Sequence hotspot (frozen tissue) | 4 of 5 ER+PR+ |
| Total | 78/2062 | 3.8% |
COSMIC database (Catalog of Somatic Mutations in Cancer; http://www.sanger.ac.uk/genetics/CGP/cosmic/) lists 57 mutations in 1618 tested breast carcinomas as of
10/30/11 (overall 3.5% mutation), including all of the above except Lauring 2010, Kalinsky 2009, Boyault 2011.
SSCP-Single strand conformation polymorphism
ER-Estrogen receptor, PR-Progesterone receptor
Table 2: Literature review of AKT1 mutations in primary invasive breast carcinomas.
| Gene | Name | Fold change Loi | Fold Change Cizkova |
|---|---|---|---|
| HMGCS2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | +2.23 | +5.31 |
| ID4 | Inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | -1.36 | +3.03 |
| LTF | Lactotransferrin | +3.23 | +10.52 |
| MSX2 | Mshhomeo box homolog 2 (Drosophila) | +1.37 | +2.17 |
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | +1.34 | +2.45 |
| WNT5A | Wingless-type MMTV integration site family, member 5A | +1.57 | +3.43 |
Table 3: Comparison of PIK3CA mutation highly up or down regulated genes: Loi supplementary dataset S1 (280 genes)[57]; Cizkova 29 gene set (Tables 3-4) [58].
In summary, current consensus associates PIK3CA mutation with favorable breast cancer outcome in ER+ tumors.
PIK3CA heterogeneity in tumors and metastasis
Several studies to date have examined PIK3CA mutational status within primary breast tumors, and between primary tumors and nodal or distant metastasis, with contrasting findings (Table 4) [74-77]. Dupont-Jensen reported considerable discordance in PIK3CA mutational status between primary tumors and metastasis [75]. In a study of 104 patients with available tissue from primary and asynchronous recurrent/metastatic carcinomas, PIK3CA mutations were found in 45% of primary tumors, with a very sensitive SNaPshot PCR method [75]. Interestingly, the rate of PIK3CA mutation in metastatic tumor was 53%, but 21 patients changed genotype from wild type to mutant, and 11 from mutant to wildtype [75]. Further, positive lymph nodes (synchronous with primary breast carcinoma) were assayed in 47 patients, revealing a 34% mutation rate, with mutational discordance between primary tumor and synchronous nodal metastasis in 12/46 patients (26%), with only 4 of 12 gaining mutation in nodal metastatic tumor [75]. Further, intratumoral heterogeneity was assayed in 10 primary tumors, and discordance was found in 4 (40%), including one case that had different foci assayed as PIK3CA wildtype, H1047R mutation, and E542K mutation (Table 4) [75].
| Author | % PIK3CA mutation invasive | % PIK3CA mutation DCIS | Concordance DCIS-invasive | Concordance Intratumoral (multiple samples from primary) | Concordance primary tumor-metastasis |
|---|---|---|---|---|---|
| Lee 2005 [51] | 31% (n=78) | 13% (n=15) | ND | ND | ND |
| Maruyama 2007 [63] | 30% (n=176) | 0% (n=12) | ND | ND | ND |
| Li 2010 [64] | 27% (n=108) | 24% (n=57) | ND | ND | ND |
| Dunlap 2010 [79] | 15% (n=81) | ND | 100% (n=13) | ND | ND |
| Miron 2010 (pilot cohort) [80] | 11% (n=34) | 19% (n=37) | 93% (n=29) | ND | ND |
| Miron 2010 (Korean cohort) [80] | 29% (n=374) | 30% (n=97) | 66% (n=45) | ND | ND |
| Da Silva 2010 [74] | 17% (n=12) | ND | ND | ND | 100% (brain n=12) |
| Dupont-Jensen 2011 [75] | 45% (n=101) | ND | ND | 60% (n=10) | 74% (nodes n=46)# 68% (distant n=100)# |
| Gonzalez-Angulo 2011 [76] | 40% (n=47) | ND | ND | ND | 82% (distant n=51)* |
| Kalinsky 2011[77] | 38% (n=63) | ND | 100% (n=5) | 95% (n=63) | 91.7% (nodes n=12)^ 100% (distant n=5) |
ND: not done
#Nodes: 8 cases had mutation in primary but not in nodal metastasis (3 exon 9, 1 exon 20); 4 had mutation in nodal metastasis but not in primary (3 exon 9, 5 exon
20).
#Distant: 11 cases had mutation in primary but not in metastasis; 21 cases had mutation in metastasis but not in primary (p=0.08)
*5 cases had mutation in primary but not in metastasis; 4 cases had mutation in metastasis but not primary
^Nodes: one case had exon 20 mutationin primary, but not in nodal metastasis
Table 4: PIK3CA mutations in DCIS, paired invasive carcinoma, and paired metastatic tumors.
Using a sensitive PCR-mass-array mutation screen, Gonzalez- Angulo et al. [76] screened another cohort of patients with metastatic breast carcinoma and found comparable PIK3CA mutation frequencies in primary tumor and metastasis after anthracycline and/or taxane based chemotherapy (40% and 42% respectively), with a propensity for exon 20 mutations in this group. However, upon analysis of paired metastasis from the same patient, 18% of patients had discordant PIK3CA mutation status. In this study, almost equal numbers of metastases gained PIK3CA mutations as lost mutations (Table 4) [76]. This group also compared wildtype and mutant peak height as a surrogate measure of the fraction of DNA/fraction of cells harboring mutation, and noted that 5 additional cases had >50% changes in the fraction of mutated DNA. While different mixtures tumor clones with heterogeneous mutation status could explain this observation, these results might also be accounted for by differing proportions of admixed non-neoplastic tissue (desmoplastic stroma etc.); although the authors attempted to control for the latter by requiring at least 70% tumor nuclear cellularity in tested specimens [76].
The group from Memorial-Sloan Kettering employed similar sensitive methodologies (PCR and mass array analysis, but also confirmed patient identity between samples by SNP analysis), with an overall mutation rate of 38%, but found substantially higher intratumoral and primary-metastasis mutational concordance. Intratumoral heterogeneity was analyzed in a larger number of primary breast cancers, with mutational concordance in 60/63 of tumors (95% see below for DCIS) [77]. Further, mutations in primary breast tumor and synchronous nodal metastasis, as well as asynchronous distant metastasis were mostly concordant (11/12, 91% nodes; 5/5, 100% distant, Table 4) [77]. In a small sample of breast carcinomas and paired brain metastases, Da Silva et al. [74] similarly found a complete concordance for PIK3CA mutation in all 12 pairs (2 pairs mutant, 10 wildtype).
The explanation for the marked difference in findings amongst these studies is unclear, perhaps related to methodological differences, divergent populations and treatments, and small samples sizes in some studies. However, a pattern is emerging, suggesting that PIK3CA mutations are neither selected for nor against in metastatic breast cancer. Further work to characterize intratumoral heterogeneity, and heterogeneity in metastatic tumors will be essential to recommending an optimal tumor testing algorithm, both for PIK3CA testing in breast carcinoma, and as applied to other types of tumors.
AKT1 mutations in breast carcinoma
Fewer studies to date have investigated AKT1 plekstrin homology domain mutations in breast cancer. Meta-analysis of 10 studies covering over 2000 samples reveals an average mutation rate of 3.8% (range 1.4-8.2%, Table 2). Interestingly, AKT1 mutations have been found almost exclusively in ER+ tumors (Table 2). Correlation with outcome has not been established, and intratumoral heterogeneity has not been investigated.
PIK3CA/AKT pathway mutations in carcinoma in-situ
Data from endometrial and colon carcinomas suggested that PIK3CA mutation may be a relatively late event in carcinogenesis [1,6,78]. Several studies have addressed the question of PIK3CA and AKT1 mutations in carcinoma in situ in the breast (Table 4), and have generally found the mutation frequency to be equivalent in ductal carcinoma in situ (DCIS) and invasive carcinoma [51,63,64,77,79,80]. Li et al. [64] reported mutations in 24% of DCIS cases (n=57), as compared to 27% of invasive carcinomas (n=108). Additionally, there is relatively good mutational concordance between DCIS and accompanying invasive carcinoma in the same specimen. Our group initially analyzed carcinoma in situ nearby invasive carcinomas which had PIK3CA or AKT1 mutations, and found 100% concordance in 13 cases, including 2 cases with AKT1 mutation in DCIS and invasive carcinoma, but we did not comprehensively examine pairs without mutated invasive carcinoma [79]. In addition to studying intratumoral heterogeneity, as discussed above, Kalinsky et al. [77] compared mutational status in 5 cases of paired DCIS and invasive carcinoma, and found complete concordance (all harboring mutations).
The largest study to date found similar PIK3CA mutation frequency among DCIS alone (30%), DCIS adjacent to invasive carcinoma (31%), DCIS paired with invasive carcinoma (DCIS=29%, IDC=31%) and invasive carcinoma alone (28%) [80]. Concordance between paired DCIS and invasive carcinoma in the group of pilot cases (which overall had a lower mutation frequency than listed above) was 93%, excluding 2 heterogeneous cases. In these two cases, multiple areas of DCIS and invasive carcinoma were tested, revealing mutational heterogeneity. In one case, 2 of 3 areas of in situ carcinoma were PIK3CA mutant, while the invasive component and one area of DCIS was wild type; in a second case, DCIS and one invasive sample was wild type, while the second invasive sample had a PIK3CA exon 20 H1047R mutation [80]. Their larger cohort demonstrated a lower DCIS-invasive mutational concordance of 66% [80]. Interestingly, discordant cases included 7 cases with mutant DCIS (6 exon 20, 1 exon 9) with accompanying PIK3CA wild type invasive carcinoma, and 8 cases with wildtype DCIS and PIK3CA mutant invasive carcinoma (all exon 20); 6 cases were concordant mutant (all exon 20), and 24 cases were concordant wildtype [80]. This data suggests that PIK3CA mutations in breast neoplasia often exist already at the stage of DCIS.
PIK3CA/AKT pathway mutations in proliferative breast lesions
Given the finding of numerous PIK3CA/AKT pathway mutations in DCIS, it is interesting to speculate whether mutations might be present in ‘earlier’ precursor or proliferative breast lesions. In addition to testing DCIS, Li et al. [64] also studied usual ductal hyperplasia (UDH), flat epithelial atypia (termed DIN 1A in their publication) and atypical ductal hyperplasia (termed DIN 1B), isolated by lasercapture microdissection and direct sequencing. They reported PIK3CA mutations in 0/16 cases of UDH, 1/20 (5%) of DIN 1A and 2/32 (6%) of DIN 1B [64].
Although our group has not studied the identical spectrum of breast lesions, our data on a variety of breast lesions suggests that PIK3CA/AKT1 mutations may be very prevalent in proliferative breast epithelium. We first studied a group of papillary neoplasms, including benign papillomas (with or without usual ductal hyperplasia), papillomas with atypia or DCIS, and papillary carcinomas, using sensitive HPLC, or PCR-mass array based methods, with sequence confirmation [81]. We found a 66% frequency of PIK3CA and AKT1 mutations in benign papillomas [81]. Interestingly, there was a bias toward AKT1 mutations in papillomas without hyperplasia, and a bias toward PIK3CA exon 20 mutations in papillomas with usual hyperplasia. Although the rate of mutation was similar in papillomas with atypia or DCIS (11/18, 61% plus a RAS mutation), the mutation rate was lower in a small sample of papillary carcinomas, and seemingly closer to that of invasive breast carcinomas (3/10, 30% plus a RAS mutation) [81].
Our group has also analyzed columnar cell lesions (with and without atypia n=24), revealing a similarly high PIK3CA mutation frequency (54% using sensitive PCR mass-array screening methods). Of the columnar cell lesions with paired DCIS or invasive carcinoma available for analysis, concordance was only 5/14 cases. Of the discordant cases, most had PIK3CA mutant columnar cell lesion, with paired wildtype carcinoma. Preliminary data from a number of additional cases is proving confirmatory, suggesting that PIK3CA mutations may be beneficial in early breast proliferative lesions, and may have a different, perhaps lesser, role in invasive and /or metastatic carcinoma. It is unclear why our results differ from Li’s group, although methodologic and target lesional differences are logical explanations.
PIK3CA mutations: Implications for drug therapy
The PI3K signaling pathway is an attractive target for pharmacologic manipulation in cancer therapy, especially in breast cancer, since it is so commonly activated. This topic has been extensively reviewed elsewhere, and will covered briefly herein [4,7,8,14,16-18,20,21]. It has been established that PIK3CA hotspot mutant isoforms do respond to inhibitors of wildtype PIK3CA, such as LY294002, wortmannin, as well as to novel pharmacologics in development [7,8,14,16-18,20,21,30,31]. A yeast screen has identified potential PIK3CA drug resistance mutations, as well as drug-sensitizing mutations [82]. However, there do not appear to be any PIK3CA mutant-isoform specific agents in development [8,16-18]. Pharmacologic agents undergoing trials include PI3K inhibitors (XL147, PX866, GDC0941, BKM120), and dual PI3K and mTOR inhibitors (BEZ235, BGT226, XL765, SF1126, GSK1059615) [8,14,16-18,20]. AKT inhibitors may be non-specific or isoform specific, and there is some data that the AKT1 E17K mutation may have differential sensitivity to plekstrin homology domain agents (AKT inhibitors include Perifosine, GSK690693, VQD002, MK2206) [8,16,18,20]. Rapamycin and its analogues are well known and generally well tolerated mTOR inhibitors, yet feedback circuits may result in AKT activation with rapamycin analogue treatment; mechanisms are still being investigated [15,17,18,83]. mTOR catalytic site inhibitors are also in the pipeline (OSI027, AZD8055) [8,18]. Inhibitors of these varying nodes along the PI3K signaling pathway will have different impact upon crosstalk and complex feedback regulatory pathways; it remains to be determined which agent or combination of agents will provide the best clinical efficacy. Furthermore, this will vary considerably between tumors based on their genotypic and phenotypic profile, including intrinsic subtype, estrogen receptor status, and Her- 2/neu status.
PI3K pathway activation has been proposed as a likely mechanism of clinical resistance to trastuzumab and lapatinib, which are agents targeted to tyrosine kinases ‘upstream’ of PI3K (Her-2/neu, dual Her-2/neu and EGFR, respectively) [12,84-88]. This is an important clinical question, as between 15-30% of Her-2neu positive tumors harbor PIK3CA mutation [46,56,76]. In vitro and translational studies have shown only weak correlation, or lack of correlation of PIK3CA mutation and drug resistance, with PTEN loss having a somewhat greater impact [12,16,84-90]. However, ‘PIK3 pathway activation,’ as indicated by either PIK3CA mutation or PTEN loss, tends to correlate with drug resistance [12,16,84-90] (it is worth re-emphasizing that activation of intracellular signaling resulting from PIK3CA mutation is different than that resulting from PTEN loss). Interestingly, O’Brien et al. demonstrated that 2 cell lines with PIK3CA H1047R mutations were lapatinib sensitive, while 2 lines with PIK3CA E545K mutation were lapatinib resistant.[85] The retrospective clinical studies to date have included predominantly heavily pretreated patients with metastatic disease [12,84,86,87]; thus, there is a great need for prospective drug studies to include thorough tumor characterization in order to address these important questions.
Conclusion
Mutations of the PI3K110α catalytic subunit (PIK3CA) are very common in estrogen receptor positive and Her-2/neu positive breast cancers, and they provide an attractive target for therapy. Since the common hotspot mutations are kinase activating, it has been hypothesized that PIK3CA/AKT1 mutations play a role in breast carcinogenesis, yet these mutations paradoxically correlate with good prognosis in estrogen receptor positive tumors. Further, several lines of data from human and mouse suggest that these mutations may have a role early in breast epithelial cell proliferation, but may have a lesser role at later stages of carcinoma and metastasis. In patient tissue, PIK3CA mutations are found in found in proliferative lesions, and in DCIS, yet metastatic tumors gain and lose PIK3CA mutation at about equal frequency, suggesting lack of selective advantage in the metastatic process. Further, mammary overexpression of PIK3CA H1047R mutant results in proliferative lesions and carcinomas, yet in the inducible mouse model, carcinoma readily lose ‘addiction’ to the PIK3CA mutant as tumors accumulate other genetic changes. Indeed, data from whole genome sequencing of human breast cancers and metastases reveals the complexity of and heterogeneity of genetic changes, with 30-90 point mutations per tumor, non-withstanding indels, translocations, epigenetic and translational/post-translational changes.[91-95] Thus, while our current knowledge of the PI3K/ AKT signalling pathway and PIK3CA/AKT1 mutations provides insight into the pathogenetics of breast cancer, further experimental studies exploring pathway crosstalk and clinical trials of combination therapies utilizing PI3K/AKT inhibitors together with other agents will be necessary to develop effective targeted treatment strategies.
Acknowledgements
Dr. Troxell is supported by a Career Catalyst grant from grant from Susan G. Komen for the Cure® (KG100112). The author would like to acknowledge the expertise and enthusiasm of collaborators in the Corless/Heinrich laboratories at OHSU, the West/van de Rijn laboratories at Stanford, and the Department of Pathology at OHSU. Alex Bolinder provided assistance with Figures 1 & 2.
References
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304: 554.
- Liu H, Radisky DC, Wang F, Bissell MJ (2004) Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol 164: 603-612.
- Jia S, Roberts TM, Zhao JJ (2009) Should individual PI3 kinase isoforms be targeted in cancer? Curr Opin Cell Biol 21: 199-208.
- Karakas B, Bachman KE, Park BH (2006) Mutation of the PIK3CA oncogene in human cancers. Br J Cancer 94: 455-459.
- Zhao L, Vogt PK (2008) Class I PI3K in oncogenic cellular transformation. Oncogene 27: 5486-5496.
- Wymann MP, Marone R (2005) Phosphoinositide 3-kinase in disease: timing, location, and scaffolding. Curr Opin Cell Biol 17: 141-149.
- McAuliffe PF, Meric-Bernstam F, Mills GB, Gonzalez-Angulo AM (2010) Deciphering the role of PI3K/Akt/mTOR pathway in breast cancer biology and pathogenesis. Clin Breast Cancer 10 Suppl 3: S59-65.
- Engelman JA (2009) Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9: 550-562.
- Wu G, Xing M, Mambo E, Huang X, Liu J, et al. (2005) Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res 7: R609-616.
- Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, et al. (2009) Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res 69: 7357-7365.
- López-Knowles E, O'Toole SA, McNeil CM, Millar EK, Qiu MR, et al. (2010) PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer 126: 1121-1131.
- Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, et al. (2010) PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol 177: 1647-1656.
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, et al. (2009) Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell 16: 463-474.
- Myers AP, Cantley LC (2010) Targeting a common collaborator in cancer development. Sci Transl Med 2: 48ps45.
- Shaw RJ, Cantley LC (2006) Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441: 424-430.
- Yuan TL, Cantley LC (2008) PI3K pathway alterations in cancer: variations on a theme. Oncogene 27: 5497-5510.
- Carracedo A, Pandolfi PP (2008) The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene 27: 5527-5541.
- Garcia-Echeverria C, Sellers WR (2008) Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene 27: 5511-5526.
- Di Cosimo S, Baselga J (2009) Phosphoinositide 3-kinase mutations in breast cancer: a "good" activating mutation? Clin Cancer Res 15: 5017-5019.
- LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA (2008) Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat 11: 32-50.
- Engelman JA, Luo J, Cantley LC (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7: 606-619.
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, et al. (2009) AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 16: 21-32.
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, et al. (2011) AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19: 58-71.
- Samuels Y, Velculescu VE (2004) Oncogenic mutations of PIK3CA in human cancers. Cell Cycle 3: 1221-1224.
- Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, et al. (2005) Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 65: 4562-4567.
- Kang S, Bader AG, Vogt PK (2005) Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A 102: 802-807.
- Bader AG, Kang S, Vogt PK (2006) Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A 103: 1475-1479.
- Vogt PK, Kang S, Elsliger MA, Gymnopoulos M (2007) Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem Sci 32: 342-349.
- Gymnopoulos M, Elsliger MA, Vogt PK (2007) Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A 104: 5569-5574.
- Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, et al. (2005) Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res 65: 10992-11000.
- Zhang H, Liu G, Dziubinski M, Yang Z, Ethier SP, et al. (2008) Comprehensive analysis of oncogenic effects of PIK3CA mutations in human mammary epithelial cells. Breast Cancer Res Treat 112: 217-227.
- Zhao L, Vogt PK (2008) Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 105: 2652-2657.
- Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, et al. (2009) Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A 106: 2835-2840.
- Lauring J, Cosgrove DP, Fontana S, Gustin JP, Konishi H, et al. (2010) Knock in of the AKT1 E17K mutation in human breast epithelial cells does not recapitulate oncogenic PIK3CA mutations. Oncogene 29: 2337-2345.
- Zhao L, Vogt PK (2010) Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 9: 596-600.
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, et al. (2007) A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448: 439-444.
- Hers I, Vincent EE, Tavaré JM (2011) Akt signalling in health and disease. Cell Signal 23: 1515-1527.
- Tokunaga E, Oki E, Egashira A, Sadanaga N, Morita M, et al. (2008) Deregulation of the Akt pathway in human cancer. Curr Cancer Drug Targets 8: 27-36.
- Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, et al. (2011) Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res 71: 2706-2717.
- Meyer DS, Brinkhaus H, Müller U, Müller M, Cardiff RD, et al. (2011) Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res 71: 4344-4351.
- Liu P, Cheng H, Santiago S, Raeder M, Zhang F, et al. (2011) Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med 17: 1116-1120.
- Borowsky A (2007) Special considerations in mouse models of breast cancer. Breast Dis 28: 29-38.
- Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, et al. (2008) Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature 453: 662-666.
- Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, et al. (2002) A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J 16: 283-292.
- Wickenden JA, Watson CJ (2010) Key signalling nodes in mammary gland development and cancer. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res 12: 202.
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, et al. (2009) Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116-4124.
- Nandi S, Guzman RC, Yang J (1995) Hormones and mammary carcinogenesis in mice, rats, and humans: a unifying hypothesis. Proc Natl Acad Sci U S A 92: 3650-3657.
- Cardiff RD, Wellings SR (1999) The comparative pathology of human and mouse mammary glands. J Mammary Gland Biol Neoplasia 4: 105-122.
- Cardiff RD (2010) The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia 15: 225-233.
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, et al. (2010) Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466: 869-873.
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, et al. (2005) PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 24: 1477-1480.
- Li SY, Rong M, Grieu F, Iacopetta B (2006) PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat 96: 91-95.
- Liang X, Lau QC, Salto-Tellez M, Putti TC, Loh M, et al. (2006) Mutational hotspot in exon 20 of PIK3CA in breast cancer among Singapore Chinese. Cancer Biol Ther 5: 544-548.
- Saal LH, Holm K, Maurer M, Memeo L, Su T, et al. (2005) PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65: 2554-2559.
- Pérez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjöld B, et al. (2007) PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res 13: 3577-3584.
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, et al. (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68: 6084-6091.
- Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, et al. (2010) PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci U S A 107: 10208-10213.
- Cizkova M, Cizeron-Clairac G, Vacher S, Susini A, Andrieu C, et al. (2010) Gene expression profiling reveals new aspects of PIK3CA mutation in ERalpha-positive breast cancer: major implication of the Wnt signaling pathway. PLoS One 5: e15647.
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, et al. (2004) The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 3: 772-775.
- Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, et al. (2004) Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 64: 7678-7681.
- Buttitta F, Felicioni L, Barassi F, Martella C, Paolizzi D, et al. (2006) PIK3CA mutation and histological type in breast carcinoma: high frequency of mutations in lobular carcinoma. J Pathol 208: 350-355.
- Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, et al. (2007) Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res 13: 6064-6069.
- Maruyama N, Miyoshi Y, Taguchi T, Tamaki Y, Monden M, et al. (2007) Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res 13: 408-414.
- Li H, Zhu R, Wang L, Zhu T, Li Q, et al. (2010) PIK3CA mutations mostly begin to develop in ductal carcinoma of the breast. Exp Mol Pathol 88: 150-155.
- Vorkas PA, Poumpouridou N, Agelaki S, Kroupis C, Georgoulias V, et al. (2010) PIK3CA hotspot mutation scanning by a novel and highly sensitive high-resolution small amplicon melting analysis method. J Mol Diagn 12: 697-704.
- Duprez R, Wilkerson PM, Lacroix-Triki M, Lambros MB, Mackay A, et al. (2012) Immunophenotypic and genomic characterization of papillary carcinomas of the breast. J Pathol 226: 427-441.
- Bleeker FE, Felicioni L, Buttitta F, Lamba S, Cardone L, et al. (2008) AKT1(E17K) in human solid tumours. Oncogene 27: 5648-5650.
- Michelucci A, Di Cristofano C, Lami A, Collecchi P, Caligo A, et al. (2009) PIK3CA in breast carcinoma: a mutational analysis of sporadic and hereditary cases. Diagn Mol Pathol 18: 200-205.
- Kehr E, Warrick A, Neff TL, Lewis R, Degnin M, et al. (2011) Invasive Mucinous Breast Carcinomas Lack PIK3CA/AKT Pathway mutations. Mod Pathol, 24: 48.
- Korcheva VB, Levine J, Beadling C, Warrick A, Countryman G, et al. (2011) Immunohistochemical and molecular markers in breast phyllodes tumors. Appl Immunohistochem Mol Morphol 19: 119-125.
- Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, et al. (2009) PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 15: 5049-5059.
- Lai YL, Mau BL, Cheng WH, Chen HM, Chiu HH, et al. (2008) PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol 15: 1064-1069.
- Ellis MJ, Lin L, Crowder R, Tao Y, Hoog J, et al. (2010) Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat 119: 379-390.
- Da Silva L, Simpson PT, Smart CE, Cocciardi S, Waddell N, et al. (2010) HER3 and downstream pathways are involved in colonization of brain metastases from breast cancer. Breast Cancer Res 12: R46.
- Dupont Jensen J, Laenkholm AV, Knoop A, Ewertz M, Bandaru R, et al. (2011) PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res 17: 667-677.
- Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K, Sahin A, Liu S, et al. (2011) PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther 10: 1093-1101.
- Kalinsky K, Heguy A, Bhanot UK, Patil S, Moynahan ME (2011) PIK3CA mutations rarely demonstrate genotypic intratumoral heterogeneity and are selected for in breast cancer progression. Breast Cancer Res Treat 129: 635-643.
- Hayes MP, Wang H, Espinal-Witter R, Douglas W, Solomon GJ, et al. (2006) PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res 12: 5932-5935.
- Dunlap J, Le C, Shukla A, Patterson J, Presnell A, et al. (2010) Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat 120: 409-418.
- Miron A, Varadi M, Carrasco D, Li H, Luongo L, et al. (2010) PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res 70: 5674-5678.
- Troxell ML, Levine J, Beadling C, Warrick A, Dunlap J, et al. (2010) High prevalence of PIK3CA/AKT pathway mutations in papillary neoplasms of the breast. Mod Pathol 23: 27-37.
- Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM (2008) Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer Cell 14: 180-192.
- Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, et al. (2011) PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 30: 2547-2557.
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, et al. (2007) A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12: 395-402.
- O'Brien NA, Browne BC, Chow L, Wang Y, Ginther C, et al. (2010) Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther 9: 1489-1502.
- Razis E, Bobos M, Kotoula V, Eleftheraki AG, Kalofonos HP, et al. (2011) Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer. Breast Cancer Res Treat 128: 447-456.
- Wang L, Zhang Q, Zhang J, Sun S, Guo H, et al. (2011) PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer 11: 11:248.
- Kataoka Y, Mukohara T, Shimada H, Saijo N, Hirai M, et al. (2010) Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann Oncol 21: 255-262.
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, et al. (2004) PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 6: 117-127.
- Fujita T, Doihara H, Kawasaki K, Takabatake D, Takahashi H, et al. (2006) PTEN activity could be a predictive marker of trastuzumab efficacy in the treatment of ErbB2-overexpressing breast cancer. Br J Cancer 94: 247-252.
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, et al. (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999-1005.
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, et al. (2009) Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809-813.
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268-274.
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153-158.
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108-1113.
- Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, et al. (2005) Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res 11: 2875-2878.
- Benvenuti S, Frattini M, Arena S, Zanon C, Cappelletti V, et al. (2008) PIK3CA cancer mutations display gender and tissue specificity patterns. Hum Mutat 29: 284-288.
- Liedtke C, Cardone L, Tordai A, Yan K, Gomez HL, et al. (2008) PIK3CA-activating mutations and chemotherapy sensitivity in stage II-III breast cancer. Breast Cancer Res 10: R27.
- Ching-Shian Leong V, Jabal MF, Leong PP, Abdullah MA, Gul YA, et al. (2008) PIK3CA gene mutations in breast carcinoma in Malaysian patients. Cancer Genet Cytogenet 187: 74-79.
- Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, et al. (2008) High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer 47: 530-542.
- Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, et al. (2010) Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat 120: 461-467.
- Bozhanov SS, Angelova SG, Krasteva ME, Markov TL, Christova SL, et al. (2010) Alterations in p53, BRCA1, ATM, PIK3CA, and HER2 genes and their effect in modifying clinicopathological characteristics and overall survival of Bulgarian patients with breast cancer. J Cancer Res Clin Oncol 136: 1657-1669.
- Boyault S, Drouet Y, Navarro C, Bachelot T, Lasset C, et al. (2011) Mutational characterization of individual breast tumors: TP53 and PI3K pathway genes are frequently and distinctively mutated in different subtypes. Breast Cancer Res Treat .
- Lerma E, Catasus L, Gallardo A, Peiro G, Alonso C, et al. (2008) Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch 453: 133-139.
- Di Nicolantonio F, Arena S, Gallicchio M, Zecchin D, Martini M, et al. (2008) Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci U S A 105: 20864-20869.
- Kim MS, Jeong EG, Yoo NJ, Lee SH (2008) Mutational analysis of oncogenic AKT E17K mutation in common solid cancers and acute leukaemias. Br J Cancer 98: 1533-1535
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 19175
- [From(publication date):
January-2012 - Dec 19, 2024] - Breakdown by view type
- HTML page views : 14532
- PDF downloads : 4643