Review Article Open Access
Parkin- An E3 Ubiquitin Ligase with Multiple Substrates
Anna Sandebring* and Angel Cedazo-Mínguez
Karolinska Institutet Department of NVS, KI-Alzheimer’s Disease Research Center, NOVUM floor 5, 141 57 Huddinge, Sweden
- Corresponding Author:
- Anna Sandebring
Karolinska Institutet., Department of NVS
KI-Alzheimer’s Disease Research Center
NOVUM, 141 57 Huddinge, Sweden
Tel: +468 58 58 36 67
Fax: +468 58 58 83 80
E-mail: anna.sandebring@ki.se
Received date: march 19, 2012; Accepted date: May 07, 2012; Published date: May 09, 2012
Citation: Sandebring A, Cedazo-MÃnguez A (2012) Parkin- An E3 Ubiquitin Ligase with Multiple Substrates. J Alzheimers Dis Parkinsonism S10:002. doi:10.4172/2161-0460.S10-002
Copyright: © 2012 Sandebring A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Alzheimers Disease & Parkinsonism
Abstract
Parkinson’s disease is a common neurodegenerative disorder. The clinical symptoms arise from a substantial loss of dopaminergic neurons in substantia nigra pars compacta, which causes motor symptoms such as bradykinesia and tremor. Although the majority of PD cases are sporadic, there is a growing number of genes shown to be involved in causing parkinsonism that manifests with similar pathology to the idiopathic disease. The most common cause to autosomal recessive parkinson’s disease (ARPD) is mutations in the gene encoding for parkin- an E3 ubiquitin ligase with widespread functions in the cell. In this review we summarize the substrates identified for parkin and which functions these imply in the cell. Elucidating the mechanism of functions of these substrates may contribute with clues on which pathways to study further in Parkinson’s disease pathology.
Abbreviations
ARPD: Autosomal Recessive Parkinson Disease; CDC-rel: Cell Division Control related protein; Drp1: Dynamin related protein 1; EGFR: Epidermal Growth Factor Receptor; FBP1: Far upstream binding element; HDAC4: Histone Deacetylase 4; Hsp70: Heat shock protein 70; IBR: in between RING; Iκκγ: Inhibitor of kappa B Kinase; KO: Knock-Out; LB: Lewy Body; Miro: Mitochondrial Rho; NF-κB: Nuclear Factor κB; Pael-R: Parkin associated endothelial receptor; PD: Parkinson Disease; PDCD2-1: Programmed cell death 2 isoform-1; PICK1: Protein Interacting with C-kinase 1; PLC: Phospholipase C; RanBP2: Ran Binding Protein 2; RING: Really Interesting New Gene; SNpc: Substantia Nigra pars compacta; TRAF2: TNF-receptor Associated Factor 2; UBL: Ubiquitin-like; VDAC: Voltage Dependent Anion Channel
Parkinson’s Disease
Parkinson’s disease (PD) is the most common neurodegenerative motor disorder and is clinically diagnosed by bradykinesia, rigidity, resting tremor and postural instability. The disease is neuropathologically characterized by substantial loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) and the presence of α-synuclein positive inclusions, termed Lewy bodies (LB) (PD reviewed in [1]).
The discovery of genes involved in the development of parkinsonism has contributed immensely to the comprehension of disease pathogenesis. Although PD is mainly a sporadic disorder, studies during the last decades have identified predisposing genetic risk factors and a direct link to 16 loci and 11 genes. A growing understanding of the genomics behind PD is providing important tools for studying disease related mechanisms. Autosomal Recessive Parkinson’s disease (ARPD) is caused by mutations in parkin, PTEN induced kinase-1 (PINK1) or DJ-1, where mutations in parkin are the most common (for reviews on genetics behind PD, see [2,3]).
Parkin is an E3 ubiquitin ligase expressed in several organs, but abundantly in the brain including the SNpc. In this review, we go through the identified parkin substrates and give an overview of which cellular functions that are associated to their respective roles, and thereby plausibly involved in PD pathogenesis.
Protein Ubiquitylation
Post-translational modification by protein ubiquitylation is the most important proteolytic quality control system in the cell. Pathological ubiquitin positive inclusion bodies in brain material from patients suffering from PD or other neurodegenerative diseases, implies that the unfolded protein response is involved in the pathogenic process ultimately leading to neuronal death [4].
The complex machinery of protein ubiquitylation engages the activity of ligases; The E1 ligases are required to activate the small ubiquitin monomers in an ATP demanding process, followed by E2 ligase conjugation to ubiquitin, which then works in conjunction with the E3 ligase to transfer ubiquitin to the E3 ligase bound substrate. The E3 ligase thereby facilitates the isopeptide bond between the substrate and ubiquitin. There are multiple shapes of ubiquitin chains, arising from the linkage between crucial lysine residues within the ubiqutin monomer. The complexity in protein ubiquitylation is thereby due to the ability of ubiqutin to generate a variety of different polymer conformations, having varying consequences for the target substrates. Ubiquitylation can lead to proteasomal degradation, but depending on the mediating ligases and the structure of the formed ubiquitin chain, targeted proteins can also undergo endocytosis and lysosomal degradation, or translocate and participate in cellular signaling (for reviews on ubiquitylation, see [5,6]). The ubiquitin pathway has been implicated in the pathogenesis of several diseases, some of them of genetic origin, including neurodegenerative diseases such as PD, ataxia and Alzheimer’s disease (for review on the ubiquitin pathway in neurodegeneration, see [7]).
Parkin is an E3 Ubiquitin Ligase
The ARPD associated gene product parkin has E3 ubiqutin ligase activity and is hence serving as a substrate recognition enzyme within the cell [8]. Parkin has an N-terminal ubiquitin-like (UBL) domain and two RING (really interesting new gene) domains, flanked by a cysteine rich in between RING (IBR) domain near the C-terlminus [9]. The RING domain E3 ligase family is the largest group of E3 ligases and is characterized by the binding of two zinc ions in a histidine and cysteine rich motif of the RING finger domain resulting in a globular conformation. The role of RING domains is to recruit ubiquitin conjugating enzymes, E2 ligases thereby bind to the E3 ligase RING domain where the ubiqutin is discharged and conjugated to the substrate [10]. Over 100 parkin mutations, including exonic rearrangements, point mutations and small deletions or insertions have been identified, which places parkin as the most common cause of ARPD [11].
Parkin mediated ubiquitylation has been shown to involve the conjugation to the E2 ubiquitin carrier proteins UbcH7 and UbcH8, which are typically involved in K48-linked polyubiquitylation in order to promote proteasomal degradation, and to UbcH13, which mediate non-degrading K63-linked polyubiquitylation [12-14]. Indeed, several of the identified parkin substrates do not accumulate in parkin knockout (KO) mice, ARPD parkin or idiopathic PD human brain, supporting the notion that parkin is able to mediate different types of ubiquitylation [15-17]. Furthermore, parkin has auto– polyubiquitylating properties, allowing the protein itself to be degraded by the proteasome [18], as well as auto– monoubiquitylation and –multiple monoubiquitylating activities in vitro [19,20]. Some of the identified mutations in the gene encoding for parkin, have been shown to impair its E3 ubiquitin ligase activity for several substrates [12,14,18]. In the sections below, we describe different cellular functions modulated by parkin via its substrates.
Synaptic Proteins and Protein Aggregation
Since PD brain pathology involves the presence of protein aggregates via an accumulation of unfolded proteins, it has been suggested that parkin has a role in avoiding the formation of such complexes by ubiquitin mediated proteasomal degradation. In line with the idea that parkin influence the amount of protein aggregation in the cell, the molecular chaperone heat-shock protein 70 (Hsp70) is regulated through parkin mediated mono-ubiquitylation [16]. Consistent with a multiple mono-ubiquitylation of Hsp70, there was no accumulation of this substrate in the insoluble fraction in brain tissue from parkin deficient ARPD subjects. However idiopathic PD patients show increased levels of Hsp70 in the insoluble fractions and decreased levels in the soluble fraction, which leaves the possibility that Hsp70 is differently activated in sporadic PD brain compared to the healthy brain. How parkin influence Hsp70 function is not elucidated, but may influence its activity.
One of the main pathological hallmarks in PD is the presence of LB, and interestingly parkin has been shown to associate with LB and further to interact with an O-glycosylated form of α-synuclein and synphilin-1, both of which are abundant LB components [17,21]. Furthermore, parkin protects against toxicity mediated by α-synuclein over-expression [22] and promotes the formation of ubiquitin positive inclusions when inhibiting the proteasome [23]. It has thus been proposed that LB formation is a protective response to toxic insults, rather than the primary cause to neuronal cell death, and that this process requires parkin activity. This may explain why the brains of parkin ARPD patient generally lack LB pathology [24,25]. Apart from being LB components, the physiological roles of α-synuclein and synphilin-1 are not fully elucidated. However, since both proteins are enriched in presynaptic terminals, they have been suggested to play a role in synaptic function. In fact, synphilin-1 associates to synaptic vesicles and this interaction appears to be modulated by α-synuclein [26].
Another pre-synaptic protein is Protein interacting with C-kinase 1 (PICK1) which is mono-ubiquitylated by parkin [27]. PICK1 is a member of the PSD95/discs large/ZO-1 (PDZ) protein family, that regulates trafficking of proteins and mediates the assembly of large protein complexes and PICK1 itself is a presynaptic protein known to associate with channels and receptors. In line with this function, the authors show that parkin overexpression abolishes the PICK1 mediated potentiation of Acid-sensing ion channel subunit 2a, suggesting that parkin mediated mono-ubiquitylation deactivates PICK1. Knockdown of parkin however enhance the excitatory effect from this channel, which may imply that ARPD involve excitotoxicity through a lack of PICK1 regulation.
Ubiquitylation and subsequent regulation of the levels of polyglutamine proteins ataxin-2 and -3 has been associated to parkin E3 ligase activity [28,29]. Parkin protects from ataxin-2 mediated neurotoxicity and is involved also in the regulation of the normal protein levels [28,29]. When polyglutamine repeats are mutated and expanded, Purkinje neurons degenerate resulting in spinocerebellar ataxia type 2. As in PD, neurodegeneration resulting from polyglutamine repeats also involves the formation of protein inclusion. Thus, it is possible that parkin participates in the clearance of misfolded proteins not only in the PD affected regions, but also for other neurodegenerative diseases.
Vesicular Dynamics
A more prominent role for parkin in vesicle formation was presented when the synaptic vesicle-enriched septin GTPases cell division control related protein (CDCrel) -1 and -2a were identified as parkin substrates [18,30]. Parkin mediated UbcH8 dependent polyubiquitylation resulted in 26S proteasomal degradation of CDCrel-1 and CDCrel-2a and both proteins were shown to accumulate in human parkin mutant ARPD brain. Septins are important for synaptic vesicle transport, fusion and recycling [31]. CDCrel-1 has been found to inhibit vesicle exocytosis by association to syntaxin [32]. A possibility is therefore that parkin, via interaction with septins, may regulate the release of dopamine. Indeed, overexpression of CDCrel-1 in substantia nigra of rats induces dopaminergic neurodegeneration and a decline in striatal dopamine levels [33]. In Drosophila melanogaster, overexpressing the CDCrel-1 homologue septin4 induced age dependent dopaminergic neurotoxicity [34]. Parkin has also been shown to ubiquitylate and promote the degradation of misfolded dopamine transporter, which resulted in a more effective dopamine uptake [35].
In line with the idea that impaired vesicular dopamine release may be related to PD pathogenesis, yet another parkin substrate associated to vesicular trafficking and dynamics is SynaptotagminXI. Parkin ubiquitylation led to proteasomal degradation of synaptotagminXI. Accumulation of synaptotagminXI was found in the core of LB of substantia nigra sections from sporadic PD patients, where also the other parkin substrates synphilin-1, p38 and far upstream binding element (FBP1) are found to accumulate [36-39].
Regulation of Genomic Translation and the Cell Cycle
The parkin substrate p38 is a structural component of the mammalian aminoacyl t-RNA synthetase complex, which are key enzymes in the translation of the genetic code. Parkin protected from p38 overexpression induced toxicity by promoting the formation of cellular inclusions [36]. Both p38 and FBP1 were also found to accumulate in brain homogenates from parkin KO mice. FBP1 is an activator of the proto-oncogene c-myc. Ubiquitylation and proteasomal degradation of FBP1 is promoted by p38 and similarly FBP1 upregulates the expression of p38, which is toxic to cells [40]. Both proteins thus appear in the same pathogenic, parkin regulated pathway.
A ubiquitin ligase complex involving parkin together with hSel-10 and Cul1 was found to ubiquitinate and regulate the levels of the cyclin dependent kinase-2 regulatory subunit cyclin E [41]. Cyclin E levels accumulated in nigral regions from both ARPD and sporadic PD brain material and the authors further show that parkin was protective to neuronal apoptosis induced by kainate excitotoxicity. Glutamatergic neurotoxicity is a feature that may be related to PD pathology and in which cyclin E has previously been reported to play a role [42].
Associated to cdk2/cyclin E is the oncogene β-catenin, a component in the Wnt signaling pathway which promotes cell proliferation [43]. Intriguingly, parkin has been shown to associate to and regulate the levels of β-catenin [44]. Parkin KO mice accumulate β-catenin and stabilizing β-catenin in cells resulted in an increase in cyclin E and associated cell death.
The E3 SUMO ligase Ran binding protein 2 (RanBP2) regulates protein shuttling between nucleus and cytosol by localizing in the cytoplasmic filament of the nuclear pore complex. Proteasomal degradation of RanBP2 is promoted by parkin ubiquitylation. A downstream effect from parkin ubiquitylation was a decreased sumolyation of the RanBP2 substrate histone deacetylase 4 (HDAC4), which repress gene transcription by chromatin condensation [45].
Programmed cell death 2 isoform-1 (PDCD2-1) is a protein involved in cell death, inflammation and proliferation. Parkin mediates the proteasome dependent ubiquitylation of PDCD2-1 and increased level of this substrate is found in substantia nigra from ARPD and sporadic PD subjects [46].
Thus, parkin is regulating cellular proliferation by association with several important cell cycle regulatory molecules. Indeed parkin has been shown to exhibit tumour suppressor effects [23,47]. The link between cancer and parkin may also be related to the destabilization of cell proliferating mechanisms through the substrates mentioned above.
Stability of Cytoskeletal Components
A putative role for parkin is in the quality control of the cellular cytoskeleton. This is based on the finding that parkin ubiquitinates and regulates the levels of α- and β- tubulins [48]. To ensure accurate cytoskeletal dynamics, the levels of cytoskeletal components are tightly controlled in an autoregulated manner. In animal cells, a feedback mechanism regulates the stability of tubulin mRNA depending on the cellular concentration of tubulin heterodimers [49]. Whether impairment in this process is influencing disease pathogenesis in ARPD patients remains to be answered, yet parkin and α- tubulin has been shown to accumulate in the insoluble fractions from cells overexpressing α-synuclein and in LB disease brains [50].
The cytoskeletal component actin can form aggregates by the activity of cofilin. Cofilin phosphorylation is regulated by Lim Kinase 1, which is another parkin substrate [51]. Parkin ubiquitylation of Lim Kinase 1 decreases cofilin activity and is thereby stabilizing the structure of actin, reversibly Lim Kinase 1 also regulate the E3 ligase activity of parkin. The authors further show that Lim Kinase 1 forms a complex together with Hsp70, parkin and CHIP, where CHIP has a stabilizing role.
Cell Survival Related Signaling
Parkin has been shown to exert its neuroprotective capacity by activating the nuclear factor-κB (NF-κB) signaling cascade [52]. This is achieved by non-proteasomal poly-ubiquitylation of the NF-κB signaling molecules Iκκγ and TRAF2. NF-κB signaling regulate genes related to cell death, differentiation and immunity and is believed to have consequences for both neuroprotection and synaptic plasticity [53].
Parkin associated endothelial receptor (Pael-R) is a substrate for which parkin has been shown to mediate degradative poly-ubiquitylation [54]. The G-protein coupled receptor Pael-R is selectively expressed in substantia nigra, accumulates in the endoplasmic reticulum and induces unfolded protein stress in cells when parkin was inactive. Levels of Pael-R are also increased in the insoluble fractions from ARPD brain homogenates, suggesting that Pael-R is an in vivo parkin substrate. Analysis from Pael-R KO and transgenic mice suggests that the Pael-R is regulating the dopaminergic content in substantia nigra neurons. Also, neurons of Pael-R transgenic mice were more susceptible to PD related toxins-induced cell death, by a mechanism involving unfolded protein stress response [55]. Furthermore, excessive Pael-R expression in parkin KO mice induced cell death [56]. Pael-R is a homologue to endothelin receptor type B, which has been shown to regulate phospholipase (PLC) activity [43] and subsequently the mobilization of intracellular Ca2+ and facilitation of Ca2+ influxes [47]. We identified PLCγ1 as a parkin substrate [57], and showed that parkin mutations or siRNA knock-down resulted in increased PLC activity and enhanced intracellular calcium levels, sensitizing cells to toxic insults [58]. Given the role of Pael-R in the induction of PLC activity, it is possible that parkin is mediating regulatory ubiquitylation of several substrates related to the same signaling pathway. Indeed, increased cytosolic calcium level is suggested to participate in PD pathogenesis [59].
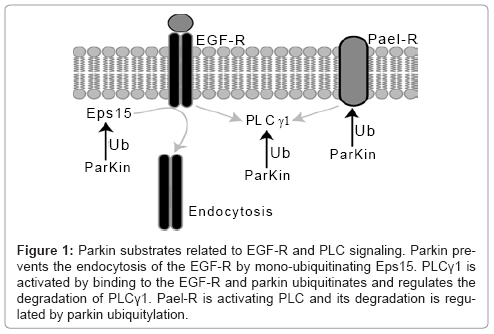
PLCγ1 is activated through binding to the epidermal growth factor receptor (EGF-R), for which internalization is mediated by another parkin substrate, epidermal growth factor receptor substrate 15 (Eps15) [60]. Parkin regulates Eps15 activity by proteasome independent mono-ubiquitylation, which results in decreased internalization of the EGF-R. EGF treatment stimulates the binding of parkin to Eps15 and to the EGF-R. Inactivation of parkin consequently results in increased EGF-R internalization and degradation and in reduced activity the prosurvival PI3K/Akt signaling pathway.
Interestingly, the parkin substrate β-catenin [44] (discussed in a section above) is downstream of the EGF mediated PI3K/Akt signaling pathway, where β-catenin is phosphorylated by pAkt regulated GSK3β [61]. Figure 1 summarizes the parkin substrates related to PLC and EGF-R signaling.
Figure 1: Parkin substrates related to EGF-R and PLC signaling. Parkin prevents the endocytosis of the EGF-R by mono-ubiquitinating Eps15. PLCγ1 is activated by binding to the EGF-R and parkin ubiquitinates and regulates the degradation of PLCγ1. Pael-R is activating PLC and its degradation is regulated by parkin ubiquitylation.
| Substrate | Physiological function | Ub. | Degradation | Ref |
|---|---|---|---|---|
| Hsp70 | Molecular chaperone | Mono | No | [16] |
| α-Sp22 | Lewy body component | Poly | Yes | [17] |
| Synphilin-1 | Interact with α-synuclein | Poly | No | [21] |
| PICK1 | Synaptic scaffolding protein | Mono | No | [27] |
| Ataxin-3 | Polyglutamine | Poly | Yes | [29] |
| Ataxin-2 | Polyglutamine | Poly | Yes | [28] |
| CDCrel-1 | Synaptic vesicle associated GTPase | Poly | Yes | [18] |
| CDCrel-2a | Synaptic vesicle associated GTPase | Poly | Yes | [30] |
| SynaptotagminXI | Membrane trafficking protein | Poly | Yes | [37] |
| p38 | Aminoacyl t-RNA synthetase cofactor | Poly | Yes | [36] |
| FBP1 | Regulates c-myc mRNA | Yes | Yes | [38] |
| Cyclin E | Cell cycle regulating protein | Poly | Yes | [41] |
| β-catenin | Component in Wnt signaling | Unknown | Yes | [44] |
| RanBP2 | Interact with nuclear pore complex | Poly | Yes | [45] |
| PDCD2-1 | Involved in apoptosis, inflammation and proliferation | Poly | Yes | [46] |
| α/β-tubulin | Cytoskeletal components | Unknown | Yes | [48] |
| Lim Kinase 1 | Phosphorylates cofilin | Poly | Yes | [51] |
| Iκκγ | Component in NFκB signaling | Poly | No | [52] |
| TRAF2 | Component in NFκB signaling | Yes | No | [52] |
| Pael-R | G-protein coupled receptor | Poly | Yes | [54] |
| PLCγ1 | Hydrolyzes lipids | Unknown | Yes | [57] |
| Eps15 | Internalizes EGF-R | Mono | No | [60] |
| VDAC | Mitochondrial ion channel | Poly | No | [63] |
| Bcl-2 | Anti-apoptotic protein | Mono | No | [15] |
| Mitofusin-1/-2 | Mitochondrial fusion protein | Poly | Yes | [70] |
| Drp1 | Mitochondrial fission protein | Poly | Yes | [71] |
| Miro | Mitochondrial anchor protein | Unknown | Yes | [72] |
Table 1: Parkin substrates (Ub: ubiquitylation by parkin).
Mitochondrial Morphology, Motility and Mitophagy
Recent studies show that parkin has a key role in the process of mitophagy- the clearance of dysfunctional mitochondria [62] by associating to the mitochondrial membrane upon toxic challenge. A couple of parkin substrates has so far been identified to be crucial for successful mitophagy; K27-linked poly-ubiquitylation of VDAC [63] and mono-ubiquitylation of the anti-apoptotic protein Bcl-2. Parkin mediated ubiquitylation of Bcl-2 enhanced Bcl-2 stability and inhibited autophagy [15]. Mitochondrial clearance is tightly connected to mitochondrial morphology, which is in turn regulated by mitochondrial fusion and fission proteins. In recent years it has become clear that all ARPD related proteins exhibits important functions for maintaining mitochondrial membrane dynamics, where PINK1 acts upstream of parkin [64-69]. When it comes to parkin substrates and mitochondrial dynamics, it was recently shown that parkin ubiquitylates the outer mitochondrial membrane fusion proteins Mitofusin-1 and -2, leading to their degradation in both a proteasomeand a AAA+ ATPase p97-dependent manner [70]. The authors further suggests that parkin mediated degradation of mitofusins is selective to dysfunctional mitochondria and thus promoting these organelles to mitophagy. Parkin also mediates the proteasomal degradation of the mitochondrial fission protein dynamin related protein-1 (Drp1) [71] where decreased parkin expression lead to increased Drp1 levels and mitochondrial fragmentation as a consequence.
A neuron specific challenge is that mitochondria and other organelles must be transported along axons in order to supply the synapse with energy. Failure of transport may be detrimental to the cell. The transport of mitochondria is obtained via the linkage to motor proteins kinesin (for anterograde transport) and dynein (for retrograde transport). Mitochondrial Rho (Miro) GTPase is together with Milton forming a complex with kinesin to link mitochondria to the microtubuli. It was lately discovered that overexpression of parkin, in concert with PINK1, could halt mitochondrial motility and this finding was further linked to parkin mediated proteasomal degradation of Miro upon mitochondrial depolarization [72]. Interestingely, Mitofusin and Miro also interact [73], which may reflect that their parkin dependent degradation are both pieces from the same puzzle.
Conclusions and Perspectives
The studies of monogenic forms of PD have generated new insights of the disease pathogenesis. The list of substrates for parkin is constantly growing and judging from their spread cellular functions, it appears that parkin has a large impact on cellular physiology. Protein misfolding and aggregates are important features in PD pathology, where the proteasome dependent polyubiquitylation mediated by parkin may be an important feature. More recently parkin has also been shown to mediate non-proteasomal ubiquitylation and is thereby regulating endocytosis or protein activity. Recent data points out similarities in functions between parkin and the other ARPD causative gene products PTEN induced kinase-1 (PINK1) and DJ-1, especially on mitochondrial morphology and mitophagy [74,75]. Some studies even suggest that parkin, PINK1 and DJ-1 form a complex [76]. Therefore it would be of interest to further study how the identified parkin substrates behave in a PINK1 or DJ-1 mutant background. Finding out if there are pathways or functions that are affected in all ARPD mutant gene products may help to dissect the more important substrates on the list and thus to identify neurodegenerative mechanisms of importance for PD which may be of value for the development of future drug targets.
Acknowledgements
This work was supported by grants from the following Swedish foundations: Riksbanken Jubileumsfonden and Parkinsonfonden.
References
- Lees AJ, Hardy J, Revesz T (2009) Parkinson's disease. Lancet 373: 2055-2066.
- Cookson MR (2010) Unravelling the role of defective genes. Prog Brain Res 183: 43-57.
- Corti O, Lesage S, Brice A (2011) What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol Rev 91: 1161-1218
- Chung KK, Dawson VL, Dawson TM (2001) The role of the ubiquitin-proteasomal pathway in Parkinson's disease and other neurodegenerative disorders. Trends Neurosci 24: S7-S14.
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425-479.
- Komander D (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937-953
- Huang Q, Figueiredo-Pereira ME (2010) Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis 15: 1292-1311.
- Matsumine H, Saito M, Shimoda-Matsubayashi S, Tanaka H, Ishikawa A, et al. (1997) Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am J Hum Genet 60: 588-596
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, et al. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605-608.
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399-434.
- Hedrich K, Eskelson C, Wilmot B, Marder K, Harris J, et al. (2004) Distribution, type, and origin of Parkin mutations: review and case studies. Mov Disord 19: 1146-1157.
- Imai Y, Soda M, Takahashi R (2000) Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem 275: 35661-35664.
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, et al. (2007) Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 178: 1025-1038.
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, et al. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25: 302-305.
- Chen D, Gao F, Li B, Wang H, Xu Y, et al. (2010) Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem 285: 38214-38223.
- Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM (2008) Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem 105: 1806-18019.
- Schlossmacher MG, Frosch MP, Gai WP, Medina M, Sharma N, et al. (2002) Parkin localizes to the Lewy bodies of Parkinson disease and dementia with Lewy bodies. Am J Pathol 160: 1655-1667.
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, et al. (2000) Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A 97: 13354-13359.
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, et al. (2006) Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem 281: 3204-3209.
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O (2006) Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet 15: 2059-2075.
- Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, et al. (2001) Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med 7: 1144-1150.
- Petrucelli L, O'Farrell C, Lockhart PJ, Baptista M, Kehoe K, et al. (2002) Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron 36: 1007-1019.
- Yang H, Zhou HY, Li B, Chen SD (2005) Neuroprotection of Parkin against apoptosis is independent of inclusion body formation. Neuroreport 16: 1117-1121.
- Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, et al. (1994) Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology 44: 437-441.
- Mori, H., et al., Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology, 1998. 51(3): p. 890-2.
- Ribeiro CS, Carneiro K, Ross CA, Menezes JR, Engelender S (2002) Synphilin-1 is developmentally localized to synaptic terminals, and its association with synaptic vesicles is modulated by alpha-synuclein. J Biol Chem 277: 23927-23933.
- Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, et al. (2007) Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell 18: 3105-3118.
- Huynh DP, Nguyen DT, Pulst-Korenberg JB, Brice A, Pulst SM (2007) Parkin is an E3 ubiquitin-ligase for normal and mutant ataxin-2 and prevents ataxin-2-induced cell death. Exp Neurol 203: 531-541.
- Tsai YC, Fishman PS, Thakor NV, Oyler GA (2003) Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem 278: 22044-22055.
- Choi P, Snyder H, Petrucelli L, Theisler C, Chong M, et al. (2003) SEPT5_v2 is a parkin-binding protein. Brain Res Mol Brain Res 117: 179-189.
- Kartmann B, Roth D (2001) Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J Cell Sci 114: 839-844.
- Beites CL, Xie H, Bowser R, Trimble WS (1999) The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci 2: 434-439.
- Dong Z, Ferger B, Paterna JC, Vogel D, Furler S, et al. (2003) Dopamine-dependent neurodegeneration in rats induced by viral vector-mediated overexpression of the parkin target protein, CDCrel-1. Proc Natl Acad Sci U S A 100: 12438-12443.
- Muñoz-Soriano V, Paricio N (2007) Overexpression of Septin 4, the Drosophila homologue of human CDCrel-1, is toxic for dopaminergic neurons. Eur J Neurosci 26: 3150-3158.
- Jiang H, Jiang Q, Feng J (2004) Parkin increases dopamine uptake by enhancing the cell surface expression of dopamine transporter. J Biol Chem 279: 54380-54386.
- Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, et al. (2003) The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet 12: 1427-1437.
- Huynh DP, Scoles DR, Nguyen D, Pulst SM (2003) The autosomal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum Mol Genet 12: 2587-2597.
- Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM (2006) Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J Biol Chem 281: 16193-16196.
- Wakabayashi K, Engelender S, Yoshimoto M, Tsuji S, Ross CA, et al. (2000) Synphilin-1 is present in Lewy bodies in Parkinson's disease. Ann Neurol 47: 521-523.
- Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, et al. (2003) Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet 34: 330-336.
- Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, et al. (2003) Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37: 735-749.
- Padmanabhan J, Park DS, Greene LA, Shelanski ML (1999) Role of cell cycle regulatory proteins in cerebellar granule neuron apoptosis. J Neurosci 19: 8747-8756.
- Ambar I, Sokolovsky M (1993) Endothelin receptors stimulate both phospholipase C and phospholipase D activities in different cell lines. Eur J Pharmacol 245: 31-41.
- Rawal N, Corti O, Sacchetti P, Ardilla-Osorio H, Sehat B, et al. (2009) Parkin protects dopaminergic neurons from excessive Wnt/beta-catenin signaling. Biochem Biophys Res Commun 388: 473-478.
- Um JW, Min DS, Rhim H, Kim J, Paik SR, et al. (2006) Parkin ubiquitinates and promotes the degradation of RanBP2. J Biol Chem 281: 3595-3603.
- Fukae J, Sato S, Shiba K, Sato K, Mori H, et al. (2009) Programmed cell death-2 isoform1 is ubiquitinated by parkin and increased in the substantia nigra of patients with autosomal recessive Parkinson's disease. FEBS Lett 583: 521-525.
- Goldman RS, Finkbeiner SM, Smith SJ (1991) Endothelin induces a sustained rise in intracellular calcium in hippocampal astrocytes. Neurosci Lett 123: 4-8.
- Ren Y, Zhao J, Feng J (2003) Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci 23: 3316-3324.
- Cleveland DW, Lopata MA, Sherline P, Kirschner MW (1981) Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell 25: 537-546.
- Kawahara K, Hashimoto M, Bar-On P, Ho GJ, Crews L, et al. (2008) alpha-Synuclein aggregates interfere with Parkin solubility and distribution: role in the pathogenesis of Parkinson disease. J Biol Chem 283: 6979-6987.
- Lim MK, Kawamura T, Ohsawa Y, Ohtsubo M, Asakawa S, et al. (2007) Parkin interacts with LIM Kinase 1 and reduces its cofilin-phosphorylation activity via ubiquitination. Exp Cell Res 313: 2858-2874.
- Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, et al. (2007) Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci 27: 1868-1878.
- Karin M, Lin A (2002) NF-kappaB at the crossroads of life and death. Nat Immunol 3: 221-227.
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, et al. (2001) An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105: 891-902.
- Imai Y, Inoue H, Kataoka A, Hua-Qin W, Masuda M, et al. (2007) Pael receptor is involved in dopamine metabolism in the nigrostriatal system. Neurosci Res 59: 413-425.
- Kitao Y, Imai Y, Ozawa K, Kataoka A, Ikeda T, et al. (2007) Pael receptor induces death of dopaminergic neurons in the substantia nigra via endoplasmic reticulum stress and dopamine toxicity, which is enhanced under condition of parkin inactivation. Hum Mol Genet 16: 50-60.
- Dehvari N, Sandebring A, Flores-Morales A, Mateos L, Chuan YC, et al. (2009) Parkin-mediated ubiquitination regulates phospholipase C-gamma1. J Cell Mol Med 13: 3061-3068.
- Sandebring A, Dehvari N, Perez-Manso M, Thomas KJ, Karpilovski E, et al. (2009) Parkin deficiency disrupts calcium homeostasis by modulating phospholipase C signalling. FEBS J 276: 5041-5052.
- Chan CS, Gertler TS, Surmeier DJ (2009) Calcium homeostasis, selective vulnerability and Parkinson's disease. Trends Neurosci 32: 249-256.
- Fallon L, Bélanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, et al. (2006) A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol 8: 834-842.
- Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411: 355-365.
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795-803.
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, et al. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119-131.
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, et al. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441: 1162-1166.
- Exner N, Treske B, Paquet D, Holmström K, Schiesling C, et al. (2007) Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci 27: 12413-12418.
- Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, et al. (2009) Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem 284: 22938-22951.
- Park J, Lee SB, Lee S, Kim Y, Song S, et al. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441: 1157-1161.
- Sandebring, A., et al., Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS One, 2009. 4(5): p. e5701.
- Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, et al. (2011) DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet 20: 40-50.
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, et al. (2010) Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191: 1367-1380.
- Wang H, Song P, Du L, Tian W, Yue W, et al. (2011) Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem 286: 11649-11658.
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, et al. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147: 893-906.
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH (2010) Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci 30: 4232-4240.
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, et al. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298.
- Dodson MW, Guo M (2007) Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson's disease. Curr Opin Neurobiol 17: 331-337.
- Xiong H, Wang D, Chen L, Choo YS, Ma H, et al. (2009) Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest 119: 650-660.
Relevant Topics
- Advanced Parkinson Treatment
- Advances in Alzheimers Therapy
- Alzheimers Medicine
- Alzheimers Products & Market Analysis
- Alzheimers Symptoms
- Degenerative Disorders
- Diagnostic Alzheimer
- Parkinson
- Parkinsonism Diagnosis
- Parkinsonism Gene Therapy
- Parkinsonism Stages and Treatment
- Stem cell Treatment Parkinson
Recommended Journals
Article Tools
Article Usage
- Total views: 14319
- [From(publication date):
specialissue-2013 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 9715
- PDF downloads : 4604