Research Article Open Access
Overexpression of the Novel Tumor Suppressor Gene FUS1 Suppresses the Growth of Small Cell Lung Cancer Cells
Cancer Cells Roza Zandi1,2*, Kai Xu1, Hans S. Poulsen2, Jack A. Roth1 and Lin Ji11Department of Thoracic and Cardiovascular Surgery, The University of Texas, M.D. Anderson Cancer Center, Houston, Texas, USA
2Department of Radiation Biology, The Finsen Centre section 6321, Copenhagen University Hospital, Copenhagen, Denmark
- *Corresponding Author:
- Roza Zandi
Department of Radiation Biology
The Finsen Centre section 6321
Copenhagen University Hospital
DK2100 Copenhagen, Denmark
Tel: 45 35456329
Fax: 45 35456301
E-mail: roza@zandi.dk, roza@rh.dk
Received Date: September 30, 2011; Accepted Date: November 25, 2011; Published Date: December 03, 2011
Citation: Zandi R, Xu K, Poulsen HS, Roth JA, Ji L (2011) Overexpression of the Novel Tumor Suppressor Gene FUS1 Suppresses the Growth of Small Cell Lung Cancer Cells. J Clinic Experiment Pathol S5:001. doi: 10.4172/2161-0681.S5-001
Copyright: © 2011 Zandi R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Abstract
FUS1, also known as tumor suppressor candidate 2 (TUSC2), is a novel candidate tumor suppressor gene (TSG) frequently inactivated in human lung cancer. Loss of FUS1 protein expression is found in almost all small cell lung cancer (SCLC) cell lines and tumor specimens. Therefore, restoration of normal FUS1 function by gene transfer could serve as a potential therapeutic strategy for the treatment of SCLC. Here we investigated the effect of exogenous expression of FUS1 by plasmid-mediated gene transfer on tumor cell growth and apoptosis induction in FUS1-defective SCLC cells. Transfection of SCLC cells with wild-type FUS1 (wt-FUS1) showed in vitro growth inhibition and a marked suppression of colony formation compared to cells transfected with an empty vector (EV) or a myristoylation-defect mutant FUS1 (mt-FUS1). Forced expression of wt-FUS1 also increased the apoptotic cell population at Sub-G0/G1 in SCLC cells compared to EV- and mt-FUS1-transfected controls, which was associated with a decreased level of pro-caspase-3 and an increased level of PARP cleavage. Our results demonstrate the potential tumor suppression function of FUS1 in SCLC cells and suggest that FUS1-mediated gene therapy could be a useful therapeutic strategy for the treatment of SCLC.
Keywords
Tumor suppressor gene; SCLC; FUS1; Apoptosis
Introduction
Small cell lung cancer (SCLC) accounts for approximately 20% of all lung cancers. It is the most aggressive of lung cancer subtypes and highly related to cigarette smoking [1]. Current treatments of SCLC are radiotherapy and chemotherapy. But despite a high rate of initial therapeutic response, relapse is common and most patients eventually succumb to the disease [1,2]. Identification of new therapeutic targets and development of novel treatment strategies are therefore urgently needed for more efficiently treating this lethal disease. One intriguing therapeutic modality is tumor suppressor gene (TSG) mediated therapy, which is based on the reintroduction of a functional TSG (therapeutic gene) into the TSG-deficient cancer cells to restore their normal gene function. Introduction of such a therapeutic gene usually leads to tumor suppression by inhibition of tumor cell proliferation and/or induction of apoptosis and other types of cell deaths in the cancer cells [3]. Although the specific genetic and molecular alterations and mechanisms leading to SCLC are still unclear, several genetic and molecular changes, including loss or inactivation of TSGs [4], have been found to play an important role in SCLC development and progression. Genetic alterations and allelic loss of the 3p chromosome, particularly in the region of 3p(14-25) [5], is frequently detected in SCLC (>90%), suggesting that the 3p arm harbors multiple TSGs [5,6]. Indeed, several candidate TSGs have been identified in the 3p region, but it remains unclear how these candidate TSGs function in suppressing SCLC development and progression [6].
FUS1, which is also known as tumor suppressor candidate 2 (TUSC2), is a novel TSG candidate that has been identified in the human chromosome 3p21.3 region [7]. Loss and reduced expression of FUS1 is seen in the majority of non-small cell lung cancer (NSCLC) and in almost all SCLC [8]. In NSCLC, loss of FUS1 expression has been associated with significantly worse overall patient survival [8]. The mechanism by which FUS1 expression is reduced or lost may include the allelic loss of the 3p21.3 chromosomal region harboring FUS1, translational repression and destabilization of the FUS1 mRNA by miRNAs, and the deficient post-translational modification of the protein [7,9-11]. Post-translational modification of FUS1 involves myristoylation of the N-terminus of the protein and is required for its tumor suppressor activity in vitro and in vivo [9]. Lack of myristoylation leads to a rapid proteasomal degradation of FUS1 protein [9]. The tumor suppressing potential of FUS1 has been studied in NSCLC cells, showing cell growth inhibition and induction of apoptosis by ectopic expression of FUS1 via plasmid- and adenoviral vector-mediated gene transfer [12,13]. Also, FUS1 knockout mice show an increased frequency of spontaneous vascular tumor development [14]. Intravenous administration of nanoparticle-encapsulated FUS1 expression vector inhibits tumor progression and metastases in NSCLC xenograft mouse models, demonstrating the promising therapeutic potential of FUS1 for the treatment of primary and disseminated human lung cancer [15]. This approach has thus been the rationale for the ongoing FUS1 nanoparticle-mediated gene therapy clinical trial for the treatment of NSCLC patients [16].
Although the tumor suppressing potential of FUS1 in NSCLC cells is well established, the function of FUS1 still remains to be investigated in SCLC. The evidence that FUS1 expression is down regulated or lost in approximately 100% of SCLCs strongly suggests that FUS1 can function as a potential tumor suppressor in SCLC. In the present study, we examined the tumor suppressor activity of FUS1 in SCLC cell lines and demonstrated that ectopic expression of wild-type (wt) FUS1 in SCLC cells by plamid gene transfer effectively inhibited the growth of SCLC by activating the apoptotic pathway. These results suggest that FUS1 may serve as a potential therapeutic gene for the treatment of SCLC.
Materials and Methods
Cell lines
The origin of all the cell lines used in this study has been described previously in detail [17,18]. SCLC cell lines DMS53 and DMS273 were maintained as monolayer cultures in Waymouth and the NSCLC cell line, H1299, in RPMI medium, respectively, with supplements of 10% fetal calf serum (FCS), 10 U/ml penicillin, and 10 μg/ml streptomycin. SCLC cell lines, GLC16 and H69, were maintained as suspension cells in RPMI medium with 10% fetal calf serum (FCS), 10U/ml penicillin, and 10μg/ml streptomycin. All tissue culture reagents were purchased from Invitrogen (Taastrup, Denmark). All cells were maintained in a humidified chamber with 5% CO2 at 37°C. All cells were tested regularly for possible mycoplasma contamination.
RT- PCR
Total human RNA from normal tissues was purchased from Clontech (Glostrup, Denmark) and Ambion (Naerum, Denmark). Total RNA from SCLC cell lines was isolated using RNA easy kit according to the manufacturer’s instruction (Qiagen). cDNA was synthesized with Superscript RT II Reverse transcriptase and amplified using Platinum TaqPolymerase (Invitrogen, Taastrup, Denmark) with 25 cycles of amplification. Primers were purchased from DNA Technology (Risskov, Denmark). Primers for FUS1, Sense: 5’- TCAGAGGCAGCAGGAGCTG, Antisence: TCACAGGGAAATCCACGTG; Primers for GAPDH, Sense: 5’-TCCATGCCATCACTGCCACCCA, Antisense: 5’-TCTTGTGCTCTTGCTGGGGCTG.
Plasmid vectors
Recombinant plasmid vectors containing the cDNA of wt FUS1 gene (wt-FUS1) and an inactive myristoylation-defect mutant FUS1 (mt-FUS1), where the predicted myristoylation site of glycine was replaced with an alanine by site-directed mutagenesis, were used in the transient transfection studies. An empty plasmid vector (EV) was used as a negative control and a green fluorescent protein (GFP) expressing plasmid vector was used to assess the transfection efficiency of the cells. All the plasmid vectors used in the experiments have a similar backbone. The construction of the recombinant plasmid vectors have been described previously [9,15].
Transient transfection
Cells were either transfected in triplicates in 96-well plates or duplicates in 6-well plates with 0,02 μg or 4 μg plasmid DNA, respectively. Adherent cells were seeded 1 day prior to transfection at concentrations varying from 10.000 to 30.000 cells/well in 96-well plate and 300.000 to 800.000 cells/well in 6-well plates, depending on the cell line used. The cells were transfected for 3 hours (h) using Lipofectamine 2000 in Opti-MEM Reduced Serum Medium (Invitrogen, Taastrup, Denmark). Suspension cells were plated at concentration of 30.000 cells/well in 96-well plate and 800.000 cells/well in 6-well plate immediately after transfection. Transfection efficiency in each cell line was assessed by parallel transfection of cells with equal amount of GFP expressing vector. Transfection efficiency was between 40-80 % in the cell lines, depending on the cell line studied. For the co-transfection studies, cells were transfected with 3.5 μg of EV, wt-FUS1 or mt-FUS plasmid vector and 0,5 μg of GFP vector per well in 6-well plates and with 0,0175 μg of EV, wt-FUS1 or mt-FUS plasmid vector and 0,0025 μg of GFP plasmid vector per well in 96-well plates, respectively.
Flow-cytometry
The cells were plated in duplicates in 6-well plates the day before or immediately after transfection with EV, wt-FUS1 and mt-FUS1 expression vectors as described above. 72h after transfection, the cells were fixed in 70% ethanol, washed with ice cold PBS and resuspended in a propidium iodide solution containing 50 μg/ml of propidium iodide, 10 mM Tris, 5 mM MgCl2, and 10 μg/ml of ribonuclease A and 1 μl/ml of NP-40 (Propidium iodide and ribonuclease A were purchased from Sigma-Aldrich, Broendby, Denmark). Samples were analyzed using Flow-cytometry (BD Biosciences, Broendby, Denmark) and data were analyzed using FACS Diva software program.
Colony formation assay
DMS273 and DMS53 cells were seeded 1 day prior to transfection at a density of 800.000 cells/well in 6-well plates. The cells were transfected with 3,5 μg of EV, wt-FUS1 or mt-FUS1 plasmid vector and 0,5 μg of pEGFP-N1 vector (Clontech, Glostrup, Denmark), which contains a neomycin/kanamycin-resistance-gene allowing for selection of transfected cells with G418. Immediately after transfection, the cells were counted and replated in dublicates in 100 mm petridishes at varied concentrations from 20.000 - 500.000 cells/dish.
DMS53 and DMS273 cells were incubated for 3-6 weeks in complete growth medium containing 1,0 mg/ml or 1,5 mg/ml G418 (Calbiochem, Herlev, Denmark), respectively. DMS273 cells grow much faster than DMS53 cells and therefore 20.000-50.000 cells/dish were sufficient to form colonies within 3 weeks. DMS53 cells, however, had to be plated at a density of 500.000 cells/dish in order to survive and be able to form colonies within 6 weeks. Colonies were fixed with 100 % ethanol, stained with Toluidine blue (Bie and Berntsen, Herlev, Denmark), and counted.
Western blot analysis
Whole cell lysates were prepared by sonication in ice-cold RIPA buffer containing 50 mM Tris-HCL, 1% NP40, 0.25% Na-deoxyholat, 150 mM NaCl, and 1 mM EDTA supplemented with Protease Inhibitor Cocktail Set III and Phosphatase Inhibitor Cocktail set II (Calbiochem, Herlev, Denmark). Proteins were resolved by SDS-PAGE and electroblotted onto nitrocellulose membranes (Invitrogen, Taastrup, Denmark). Membranes were blocked in 5% non-fat milk for 1 h and then incubated with primary antibody overnight at 4°C followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody. The HRP signal was detected using Supersignal West Dura Extended Duration Substrate (Pierce Biotechnology, Herlev, Denmark) in an Autochemi system (UVP).
The antibodies used were: Anti-PARP (Cell Signaling, Glostrup, Denmark), anti-caspase-3 (Santa Cruz, Aarhus, Denmark), anti- GAPDH (Santa Cruz, Aarhus, Denmark), anti-FUS1 polyclonal antibodies were developed as described previously [9], HRP-conjugated swine anti-rabbit (Dako, Glostrup, Denmark) and HRP-conjugated rabbit anti-mouse (Dako, Glostrup, Denmark).
Results
Ectopic expression of FUS1 inhibits the growth of SCLC cells lacking endogenous FUS1 protein
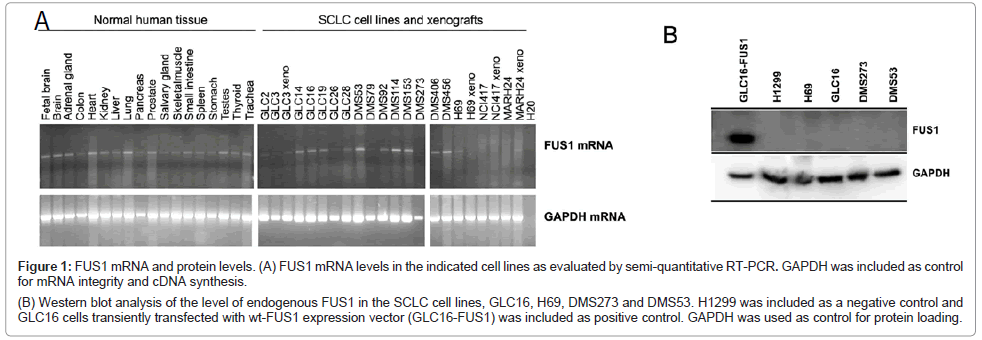
We first determined the expression of FUS1 gene in a large panel of normal tissue and SCLC cells lines by semi-quantitative RT-PCR (Figure 1A). FUS1 mRNA transcripts were detected in all the normal tissues including lung, having high levels of FUS1 gene expression. The expression of FUS1 was also detected in the majority of the SCLC cell lines with varied levels (Figure 1A). Four SCLC cell lines were chosen to further study the level of FUS1 protein expression (Figure 1B). The protein expression of FUS1 in all four SCLC cell lines was completely absent. The human NSCLC cell line, H1299, and the SCLC cell line, GLC16, that was transiently transfected with FUS1 (GLC16-FUS1) were included as negative and positive controls, respectively (Figure 1B). In addition to lacking FUS1 protein expression, the four SCLC cell lines were also chosen for this study for their relatively higher transfection efficiency with plasmid vectors than other SCLC cell lines tested (data not shown).
Figure 1: FUS1 mRNA and protein levels. (A) FUS1 mRNA levels in the indicated cell lines as evaluated by semi-quantitative RT-PCR. GAPDH was included as control for mRNA integrity and cDNA synthesis.
(B) Western blot analysis of the level of endogenous FUS1 in the SCLC cell lines, GLC16, H69, DMS273 and DMS53. H1299 was included as a negative control and
GLC16 cells transiently transfected with wt-FUS1 expression vector (GLC16-FUS1) was included as positive control. GAPDH was used as control for protein loading.
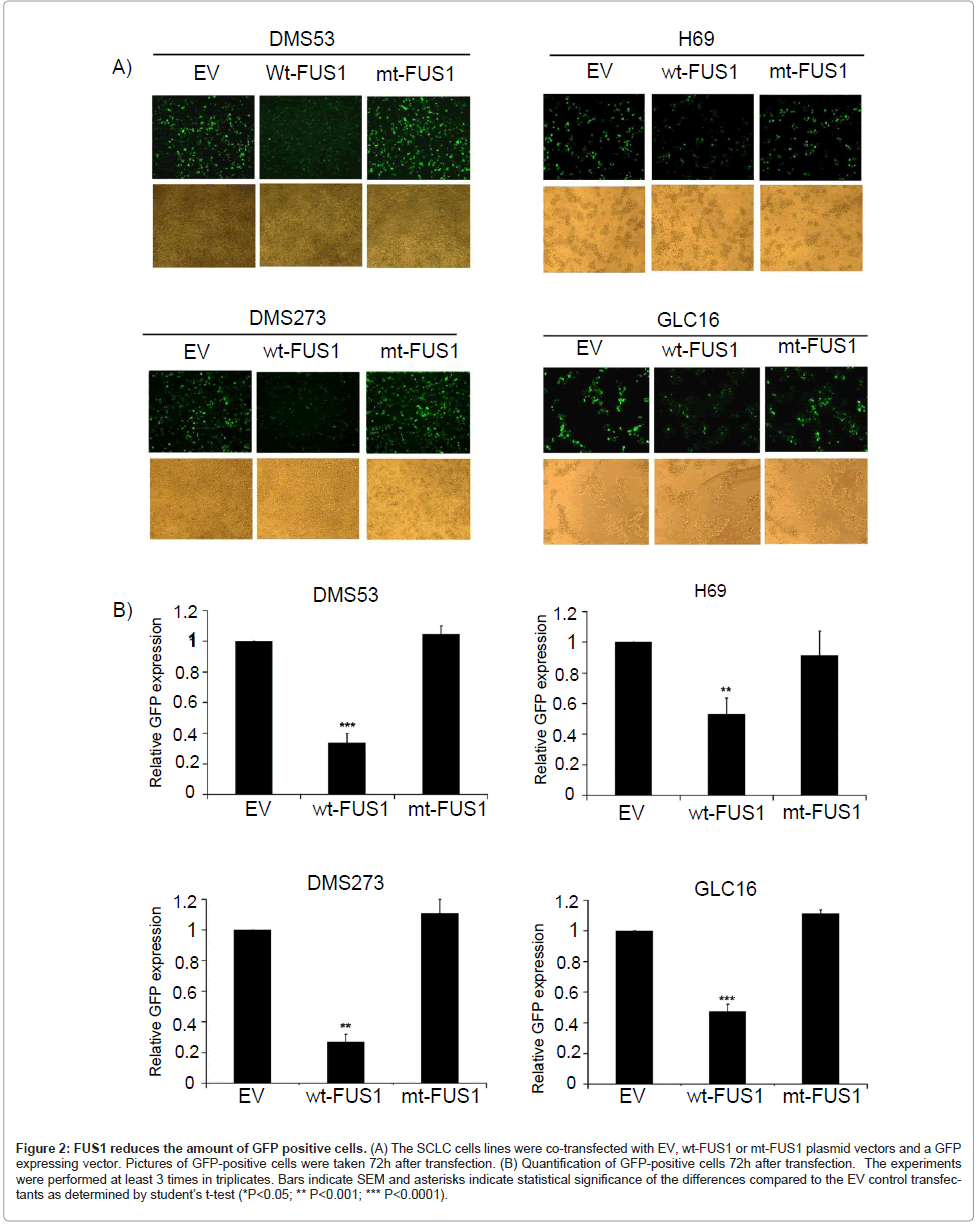
To test whether ectopic expression of FUS1 would inhibit the growth of these SCLC cells and to minimize the effects caused by the variations in the transfection efficiency in these SCLC cell lines (data not shown), we co-transfected the SCLC cells with either an expression vector containing wt FUS1 (wt-FUS1), an empty vector (EV) or a myristoylation-site-deficient mutant FUS1 (mt-FUS1) together with a GFP expressing vector. The effect of wt-FUS1 expression on tumor cell growth was evaluated by measuring the level of GFP expression in these FUS1-GFP co-transfected cells, using EV as a negative control. Because all the plasmids have the same backbone with no major size differences, the level of exogenous gene expression in the transfected cells is expected to be similar. Therefore, the level of GFP expression can be used as a convenient indicator for the measurement of cell killing by the co-transfected therapeutic genes. As shown in Figure 2A-B, a significant reduction in the GFP-positive population was observed in wt-FUS1-transfected SCLC cells compared to those in EVor mt-FUS1- transfected cells. Even though the transfection efficiency of H69 cells was low compared to DMS53, DMS273 and GLC16 cells, a significantly decreased level of GFP was detected in H69 cells transfected with wt-FUS1 compared to those transfected with EV and mt-FUS1 (Figure 2A-B). These results provide solid support for FUS1- induced tumor cell growth inhibition on these SCLC, excluding the influences of variations in transfection efficiency.
Figure 2: FUS1 reduces the amount of GFP positive cells. (A) The SCLC cells lines were co-transfected with EV, wt-FUS1 or mt-FUS1 plasmid vectors and a GFP expressing vector. Pictures of GFP-positive cells were taken 72h after transfection. (B) Quantification of GFP-positive cells 72h after transfection. The experiments were performed at least 3 times in triplicates. Bars indicate SEM and asterisks indicate statistical significance of the differences compared to the EV control transfectants as determined by student├ó┬?┬?s t-test (*P<0.05; ** P<0.001; *** P<0.0001).
FUS1 inhibits SCLC cell-induced colony formation
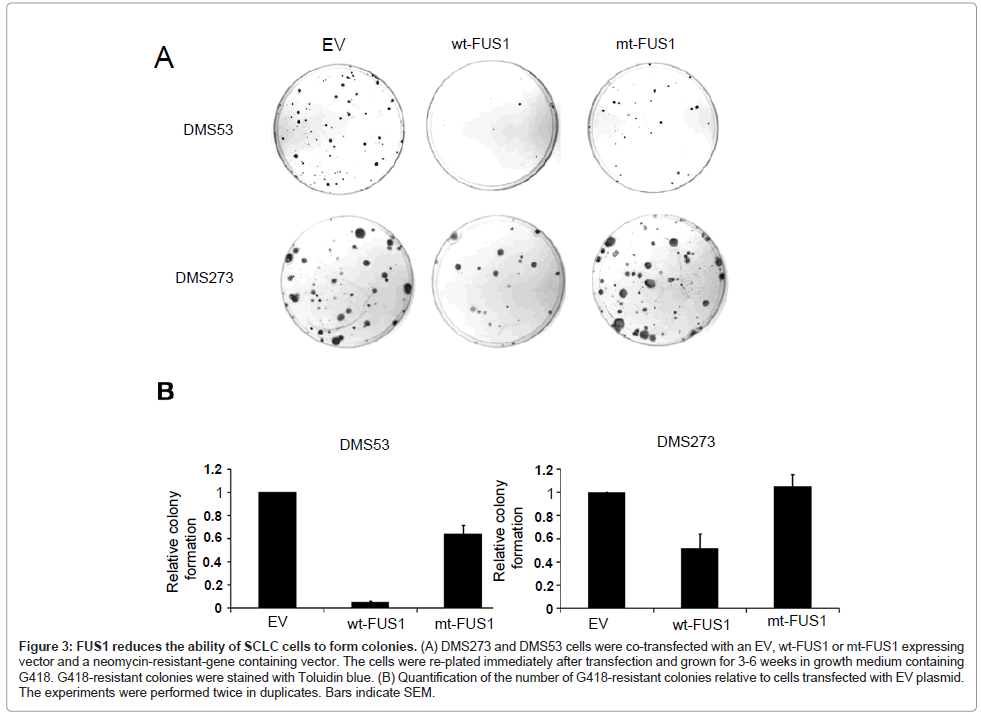
The effect of exogenous FUS1 expression on tumor cell-induced colony formation was investigated in the two adherent SCLC cell lines, DMS53 and DMS273, by colony formation assay. The cells were transiently co-transfected with EV, wt-FUS1 or mt-FUS1 and a neomycin-resistant-gene containing expression vector. The transfected cells were re-plated and selected with G418. An almost 50% reduction of colony formation was observed in the DMS273 cells transfected with wt-FUS1 compared to those transfected with EV and mt-FUS1 (Figure 3A-B). In DMS53 cells, transfection with wt-FUS1 reduced the ability of the transfected cells to form colonies by more than 90% compared to transfection with EV (Figure 3A-B). A reduced number of colonies was also observed in the DMS53 cells transfected with mt-FUS1 compared to those transfected with EV but to a much lesser extend than those transfected with wt-FUS1 (Figure 3A-B). These results confirm the growth suppressing effect of FUS1 on SCLC cells.
Figure 3: FUS1 reduces the ability of SCLC cells to form colonies. (A) DMS273 and DMS53 cells were co-transfected with an EV, wt-FUS1 or mt-FUS1 expressing vector and a neomycin-resistant-gene containing vector. The cells were re-plated immediately after transfection and grown for 3-6 weeks in growth medium containing G418. G418-resistant colonies were stained with Toluidin blue. (B) Quantification of the number of G418-resistant colonies relative to cells transfected with EV plasmid. The experiments were performed twice in duplicates. Bars indicate SEM.
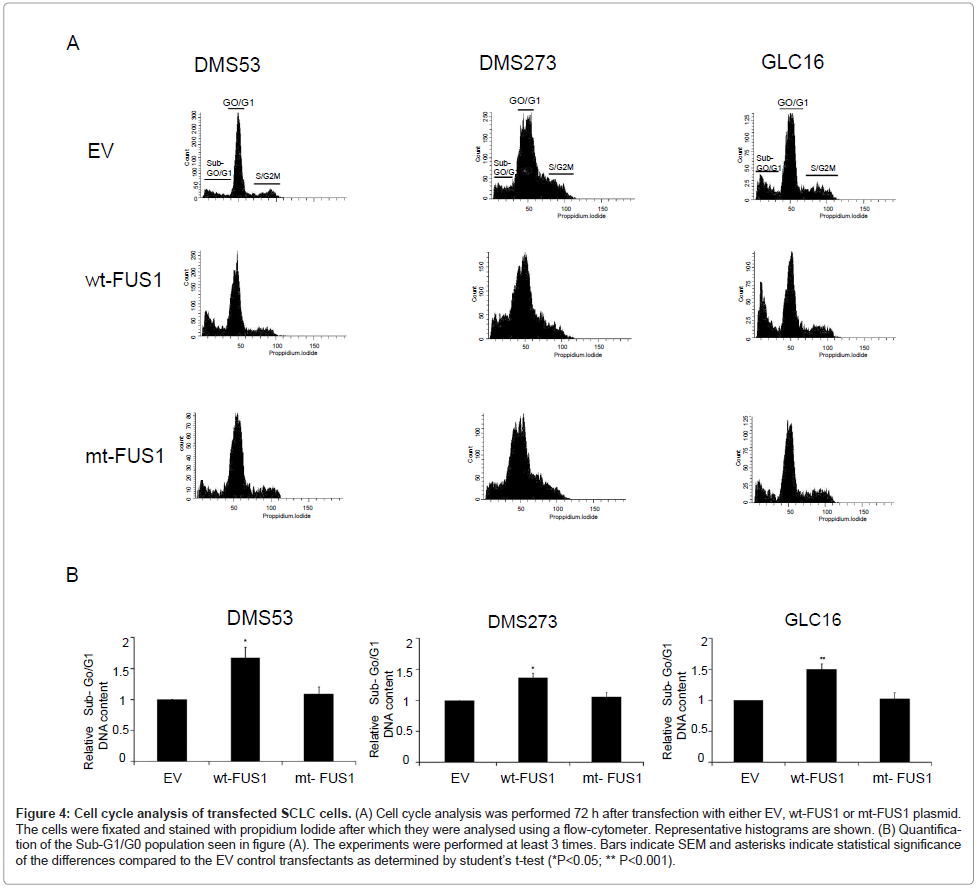
Effect of FUS1 on cell-cycle kinetics
To determine the mechanism in the above observed tumor cell growth inhibition mediated by ectopic expression of FUS1, we examined the effects of FUS1 expression on cell-cycle kinetics and apoptosis induction in the SCLC cells transfected with EV, wt-FUS1, or mt-FUS1 by FACS analysis with propidium-iodide staining. A significant increased fraction of cells with Sub-G0/G1 DNA content was detected in the SCLC cell lines, DMS53, DMS273 and GLC16, transfected with wt-FUS1 compared to those transfected with EV and mt-FUS1 (Figure 4A-B). In addition, the percentage of cells in G0/G1 and S/G2/M phase in FUS1-transfected cells decreased proportionally compared to those in EV and mt-FUS1 transfected cells in all three SCLC cell lines (Supplementary figure 1). These results suggest that FUS1 induces cell death in SCLC cells by induction of apoptosis.
Figure 4: Cell cycle analysis of transfected SCLC cells. (A) Cell cycle analysis was performed 72 h after transfection with either EV, wt-FUS1 or mt-FUS1 plasmid. The cells were fixated and stained with propidium Iodide after which they were analysed using a flow-cytometer. Representative histograms are shown. (B) Quantification of the Sub-G1/G0 population seen in figure (A). The experiments were performed at least 3 times. Bars indicate SEM and asterisks indicate statistical significance of the differences compared to the EV control transfectants as determined by student├ó┬?┬?s t-test (*P<0.05; ** P<0.001).
Overexpression of FUS1 activates pro-apoptotic proteins in SCLC cells
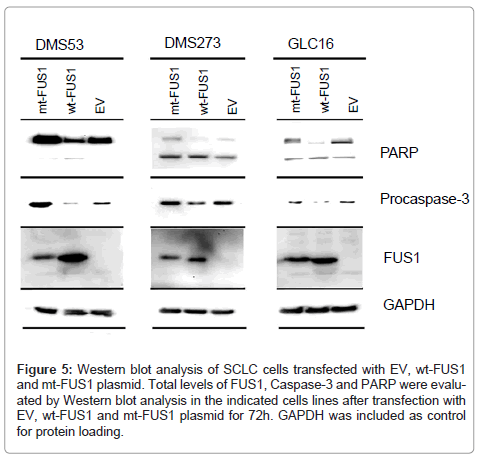
To determine if the observed FUS1-induced cell death in SCLC cells was indeed associated with FUS1-mediated activation of the apoptotic pathway, we analyzed the level of several pro-apoptotic proteins in FUS1 transfected cells by Western blot analysis. A reduced level of procaspase-3, an indication of the caspase activation by the proteolytic cleavage, was observed in the SCLC cell lines DMS53, DMS273, and GLC16, transfected with wt-FUS1 (Figure 5). This observed caspase-3 activation was correlated with an increased level of cleavage of PARP protein, which is a downstream target of caspase-3, in these wt-FUS1- transfected SCLC cells (Figure 5). Taken together, these results suggest FUS1-induced cell death is mediated by activation of the apoptotic pathway involving caspase-3 and PARP.
Figure 5: Western blot analysis of SCLC cells transfected with EV, wt-FUS1 and mt-FUS1 plasmid. Total levels of FUS1, Caspase-3 and PARP were evaluated by Western blot analysis in the indicated cells lines after transfection with EV, wt-FUS1 and mt-FUS1 plasmid for 72h. GAPDH was included as control for protein loading.
Discussion
FUS1 is a novel candidate TSG frequently inactivated in lung cancer. Loss of FUS1 expression found in almost all SCLC cell lines and tumor tissue specimen [8] suggest that FUS1 might function as a tumor suppressor in SCLC. In accordance with previous findings [7], FUS1 mRNA was detected in the majority of the SCLC cell lines and xenograft tissues. Despite the presence of FUS1 mRNA in the four SCLC cell lines, GLC16, H69, DMS53 and DMS273, no protein expression could be detected, which is in agreement with previous reports showing loss or reduced FUS1 protein expression in SCLC [8]. How FUS1 is inactivated in SCLC cells is not clear. Mutation in the FUS1 gene in lung cancer is rarely found and promoter methylation is unlikely a possible mechanism of inactivation because of the presence of FUS1 mRNA detected in these tumor cells [9,12]. Recently, Du et al. [10] reported that the expression of FUS1 is negatively regulated by selected miRNAs. Of the three miRNAs that were found to repress FUS1 protein but not mRNA expression, two (miR-93 and miR-98) were found to be highly expressed in SCLC cell lines compared to NSCLC and human bronchial cells [10]. Therefore, upregulated expression of miR93 and miR-98 could contribute to the loss of FUS1 protein expression observed in the SCLC cells. Another possible explanation could be loss of myristoylation of FUS1 protein in the SCLC cells. A myristoylation defect of the FUS1 protein has been found in primary lung cancers [9]. Myristoylation is important for FUS1 stability, as shown by the evidence that a myristoylation defective FUS1 protein is rapidly degraded through a proteasome-dependent pathway [9].
Since myristoylation of FUS1 has been shown to be required for its tumor suppressing activity [9], we compared the tumor suppression activity of the wt-FUS1 with that of the myristoylation-defect mutant FUS1 (mt-FUS1) in SCLC cells. Ectopic expression of wt-FUS1 effectively inhibited the growth of the SCLC cell lines DMS53, GLC16 and DMS273, which exhibited high plasmid transfection efficiency, but no significant inhibitory effects were detectable by expression of mt-FUS1 (data not shown). In the H69 cell line, no difference in cell viability could be observed between EV, wt-FUS1 and mt-FUS1 transfected cells, probably due to the low transfection efficiency in this cell line (data not shown). However, when H69 cells were cotransfected with GFP and EV, wt-FUS1 or mt-FUS1, a clear difference in GFP expression could be seen among these transfectants. H69 cells co-transfected with wt-FUS1 had a lower GFP expression compared to those transfected with EV and mt-FUS1. The same was observed in the other three SCLC cell lines with GFP-cotransfection. Our results suggest that the reduction in the GFP-positive population can be used as a more accurate measurement of FUS1-mediated tumor cell killing by eliminating the negative effects introduced by variations in transfection efficiency. The co-transfection method is therefore particularly useful in cells with poor transfection efficiency and where the evaluation of the cytotoxic potential of a gene is not possible with standard cytotoxicity assays as they rely on high transfection efficiency.
The FUS1-mediated inhibitory effect on the growth of SCLC cells was further supported by the FUS1-mediated reduction of colony formation in the two adherent SCLC cells lines, DMS53 and DMS273, which is consistent with previous findings in NSCLC cells [9]. Like the GFP co-transfection study, the colony formation assay was used to evaluate effects of therapeutic genes by plasmid vector-mediated gene transfer on tumor cell-induced colony formation in survival transfectants under G418 selection pressure. The comparable results that were observed in both the GFP co-transfection study and colony formation assay further supports the use of the co-transfection study as useful and more efficient method for the evaluation of the cytotoxic potential of specific genes in cells with varied transfection efficiency.
Cell cycle analysis of wt-FUS1 transfected SCLC cells showed a significant increase in the fraction of cells with Sub-G0/G1 DNA content in the DMS53, DMS273 and GLC16 cells, suggesting an increased level of apoptosis induction. The fact that the percentage of cells in G0/G1 and S/G2/M phase in FUS1-transfected cells decreased proportionally with the increase in Sub-G0/G1 population suggests that FUS1 induces cell death by apoptosis induction rather than by cell–cycle arrest in SCLC cells. To determine if FUS1-mediated SCLC growth inhibition was indeed mediated by apoptosis, we analyzed the level of several proteins involved in apoptosis and proliferation pathways by Western blot analysis. The PI3K/Akt signaling pathway has been shown to play an important role in inducing SCLC cell proliferation and survival, as inhibition of the PI3K/Akt pathway inhibits cells growth and induces cell death in SCLC [19-23]. However, in SCLC cells, ectopic expression of wt-FUS1 appeared to have no significant effect on the level of phosphorylated Akt (data not shown), suggesting that the downregulation of the PI3K/Akt signaling pathway may not be the potential mechanism in the FUS1-mediated tumor cell growth inhibition in SCLC cells. FUS1 did however reduce the total level of procaspase-3 and induce PARP cleavage, which are hallmarks of apoptosis, in the SCLC cell lines. FUS1 has been shown to induce apoptosis in NSCLC by upregulating Apaf-1 and activating the caspase cascade [25,26]. Since our results suggest that FUS1 induces cell death in SCLC cells by inducing a caspase-dependent apoptosis, it is likely that the mechanism by which it does so is similar to that seen in NSCLC.
In summary, our results demonstrate that ectopic expression of wt-FUS1 can inhibit SCLC cell growth by induction of apoptosis, providing evidence for its tumor suppressing potential in SCLC. FUS1 could therefore serve as a potential therapeutic agent for the treatment of SCLC either alone or in combination with other TSGs or therapeutic drugs. FUS1 has been shown to act synergistically with the chemotherapeutic agent cisplatin to inhibit tumor cell growth in NSCLC [25]. A similar combination treatment strategy may be used for a more effective treatment of SCLC. It also raises the possibility of using FUS1 in combination with other pro-apoptotic TSGs [27]. Therapeutic synergism between the two TSGs, FUS1 and p53, has also been demonstrated in NSCLC [26]. However, due to high levels of mutant p53 in SCLC cells, introduction of wt p53 to SCLC cells may not be effective due to dominant negative effects of the mutant protein. Instead, FUS1 can be combined with low molecular weight compounds such as PRIMA-1 or PRIMA-1Met, which are capable of reactivating mutant p53 [28-30]. Combination therapy involving FUS1 could allow a more effective treatment strategy for patients with SCLC.
Acknowledgements
This work was partially supported by grants from the National Cancer Institute, National Institutes of Health, SPORE P50CA70907 (Minna and Roth) and R01CA116322 (Ji), the A. P. Moeller Foundation for the Advancement of Medical Science, Novo Nordisk Foundation, Danish Cancer Society, Danish Medical Research Council, Aase and Ejnar Danielsen Foundation, Kathrine and Vigo Skovgaards foundation and VFK Krebsforschung gGmbH in Germany. Roza Zandi was funded by grants from the Fulbright Commission, the Dagmar Marshall foundation, the Lundbeck foundation, and the University of Copenhagen.
References
- Cooper S, Spiro SG (2006) Small cell lung cancer: Treatment review. Respirology 11: 241-248.
- Simon M, Argiris A, Murren JR (2004) Progress in the therapy of small cell lung cancer. Crit Rev Oncol Hematol 49: 119-133.
- El Aneed A (2004) Current strategies in cancer gene therapy. Eur J Pharmacol 498: 1-8.
- Sanchez-Cespedes M (2003) Dissecting the genetic alterations involved in lung carcinogenesis. Lung Cancer 40: 111-121.
- Jackman DM, Johnson BE (2005) Small-cell lung cancer. Lancet 366: 1385-1396.
- Yokota J, Kohno T (2004) Molecular footprints of human lung cancer progression. Cancer Sci 95: 197-204.
- Lerman MI, Minna JD (2000) The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res 60: 6116-6133.
- Prudkin L, Behrens C, Liu DD, Zhou X, Ozburn NC, et al. (2008) Loss and reduction of FUS1 protein expression is a frequent phenomenon in the pathogenesis of lung cancer. Clin Cancer Res 14: 41-47.
- Uno F, Sasaki J, Nishizaki M, Carboni G, Xu K, et al. (2004) Myristoylation of the fus1 protein is required for tumor suppression in human lung cancer cells. Cancer Res 64: 2969-2976.
- Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, et al. (2009) miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res 7: 1234-1243.
- Lee DY, Deng Z, Wang CH, Yang BB (2007) MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A 104: 20350-20355.
- Kondo M, Ji L, Kamibayashi C, Tomizawa Y, Randle D, et al. (2001) Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21.3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene 20: 6258-6262.
- Ji L, Nishizaki M, Gao B, Burbee D, Kondo M , et al. (2002) Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res 62: 2715-2720.
- Ivanova AV , Ivanov SV, Pascal V, Lumsden JM, Ward JM, et al. (2007) Autoimmunity, spontaneous tumourigenesis, and IL-15 insufficiency in mice with a targeted disruption of the tumour suppressor gene Fus1. J Pathol 211: 591-601.
- Ito I, Ji L, Tanaka F, Saito Y, Gopalan B, et al. (2004) Liposomal vector mediated delivery of the 3p FUS1 gene demonstrates potent antitumor activity against human lung cancer in vivo. Cancer Gene Ther 11: 733-739.
- (2003) Phase I study of IV DOTAP: Cholesterole-FUS1 in Non-Small-Cell Lung Cancer.
- Pedersen N, Mortensen S, Sorensen SB, Pedersen MW, Rieneck K, et al. (2003) Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res 63: 1943-1953.
- Poulsen TT, Pedersen N, Juel H, Poulsen HS (2008) A chimeric fusion of the hASH1 and EZH2 promoters mediates high and specific reporter and suicide gene expression and cytotoxicity in small cell lung cancer cells. Cancer Gene Ther 15: 563-575.
- Krystal GW, Sulanke G, Litz J (2002) Inhibition of phosphatidylinositol 3-kinase-Akt signaling blocks growth, promotes apoptosis, and enhances sensitivity of small cell lung cancer cells to chemotherapy. Mol Cancer Ther 1: 913-922.
- Moore SM, Rintoul RC, Walker TR, Chilvers ER, Haslett C, et al. (1998) The presence of a constitutively active phosphoinositide 3-kinase in small cell lung cancer cells mediates anchorage-independent proliferation via a protein kinase B and p70s6k-dependent pathway. Cancer Res 58: 5239-5247.
- Hodkinson PS, Elliott T, Wong WS, Rintoul RC, Mackinnon AC, et al. (2006) ECM overrides DNA damage-induced cell cycle arrest and apoptosis in small-cell lung cancer cells through beta1 integrin-dependent activation of PI3-kinase. Cell Death Differ 13: 1776-1788.
- Marinov M, Ziogas A, Pardo OE, Tan LT, Dhillon T, et al. (2009) AKT/mTOR pathway activation and BCL-2 family proteins modulate the sensitivity of human small cell lung cancer cells to RAD001. Clin Cancer Res 15: 1277-1287.
- Kraus AC, Ferber I, Bachmann SO, Specht H, Wimmel A, et al. (2002) In vitro chemo- and radio-resistance in small cell lung cancer correlates with cell adhesion and constitutive activation of AKT and MAP kinase pathways. Oncogene 21: 8683-8695.
- Ji L, Roth JA (2008) Tumor suppressor FUS1 signaling pathway. J Thorac Oncol 3: 327-330.
- Deng WG, Wu G, Ueda K, Xu K, Roth JA, et al. (2008) Enhancement of antitumor activity of cisplatin in human lung cancer cells by tumor suppressor FUS1. Cancer Gene Ther 15: 29-39.
- Deng WG, Kawashima H, Wu G, Jayachandran G, Xu K, et al. (2007) Synergistic tumor suppression by coexpression of FUS1 and p53 is associated with down-regulation of murine double minute-2 and activation of the apoptotic protease-activating factor 1-dependent apoptotic pathway in human non-small cell lung cancer cells. Cancer Res 67: 709-717.
- Christensen CL, Zandi R, Gjetting T, Cramer F, Poulsen HS (2009) Specifically targeted gene therapy for small-cell lung cancer. Expert Rev Anticancer Ther 9: 437-452.
- Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, et al. (2002) Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med 8: 282-288.
- Bykov VJ, Issaeva N, Selivanova G, Wiman KG (2002) Mutant p53-dependent growth suppression distinguishes PRIMA-1 from known anticancer drugs: a statistical analysis of information in the National Cancer Institute database. Carcinogenesis 23: 2011-2018.
- Bykov VJ, Zache N, Stridh H, Westman J, Bergman J, et al. (2005) PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene 24: 3484-3491.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15606
- [From(publication date):
December-2011 - Apr 05, 2025] - Breakdown by view type
- HTML page views : 11014
- PDF downloads : 4592