Research Article Open Access
Optimization of Fermentation Conditions of the Engineered Corynebacterium Glutamicum to Enhance L-Ornithine Production by Response Surface Methodology
Dong-Mei Lu, Ling-Yan Jiang, Lu-An Chen, Jian-Zhong Liu* and Zong-Wan MaoThe Key Laboratory of Gene Engineering of Ministry of Education and Biotechnology Research Center, Sun Yat-Sen University, Guangzhou 510275, P.R.China
- Corresponding Author:
- Jian-Zhong Liu
The Key Laboratory of Gene Engineering of Ministry of
Education and Biotechnology Research Center
Sun Yat-Sen University, Guangzhou 510275, P.R.China
Tel: 86-20-84110115
Fax: 86-20-84110115
E-mail: lssljz@mail.sysu.edu.cn
Received date: November 01, 2011; Accepted date: November 05, 2011; Published date: November 07, 2011
Citation: Lu DM, Jiang LY, Chen LA, Liu JZ, Mao ZW (2011) Optimization of Fermentation Conditions of the Engineered Corynebacterium Glutamicum to Enhance L-Ornithine Production by Response Surface Methodology. J Biotechnol Biomaterial 1:116. doi: 10.4172/2155-952X.1000116
Copyright: © 2011 Lu DM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Optimization of fermentation conditions for L-ornithine production using the engineered Corynebacterium glutamicum ATCC13032 (? argF? pro? kgd) was carried out by one-factor-at-a-time, Plackett-Burman, the steepest ascent design and a response surface methodology. Addition of Tween 80, tricarboxylic acid cycle intermediates (KAc and succinic acid) improved L-ornithine production. Using Plackett-Burman design, glucose, (NH4)2SO4, MgSO4- 7H2O and yeast extract were screened as significant factors. Response surface methodology was used to further optimize the fermentation medium. The optimized medium contained (g/L): glucose 80.00, yeast extract 13.99, (NH4)2SO4 37.94, MgSO4-7H2O 1.58, KH2PO4 1.00, K2HPO4 0.50, Na2HPO4 0.50, FeSO4-7H2O 0.02, MnSO4-4H2O 0.02, molasses 2.00, KAc 5.00, succinic acid 5.00, CaCO3 10 and Tween 80 1ml (addition after 8 h of fermentation). L-ornithine production was increased by 197 % compared to that under unoptimized conditions. 18.17 g/L of L-ornithine production was obtained at 60 h of fermentation in 5 L bioreactor by the engineered Corynebacterium glutamicum, which is the highest level in engineered strain so far.
Keywords
Optimization; Plackett-Burman Design; Response Surface Methodology; L-Ornithine; Engineered Corynebacterium Glutamicum; Bioreactor
Introduction
Corynebacterium Glutamicum (C. glutamicum) is a gram-positive soil bacterium that is widely used for the production of a variety of amino acids. The ability of C. glutamicum strains to produce certain amino acids results from several improvements made through repeated random mutations and selection [1-3]. L-ornithine is an important constituent of the urea cycle and the precursor of other amino acids such as citrulline, glutamic acid, and proline. It plays key function to discharge amino nitrogen and solve ammonia toxicity. Recently, Lornithine is widely applied in the fields of the function beverage, diet, health care, liver-protecting, anticancer, and in the food and medicine industry [4-6].
Many studies have reported that L-ornithine was produced from a citrulline- or arginine-required mutant of a coryneform bacterium obtained by classical mutagenesis [7-9]. Although this mutant can produce a high yield of L-ornithine, its growth culture is always unstable because of reversion of the auxotrophic mutant, and then the production of Lornithine drops markedly. Recently, metabolic engineering has become a powerful approach for strain improving. It may also be used to enhance the flux to L-ornithine. Only three papers reported the results of enhance of L-ornithine production by metabolic engineering so far [10-12]. Lee and Cho [10] reported that an engineered E. coli W3110 (Δ argFΔargIΔargRΔproBΔspeFParaB-arg214) produced L-ornithine of 13.2 mg/gDCW. Hwang et al. [11] also reported that cooverexpression in a triple gene knockout Corynebacterium Glutamicum (ΔargFΔargR ΔproB) resulted in L-ornithine production of 16.49 mg/g DCW, L-ornithine concentration of 179.14 mg/L. It has been reported that proline could enhance L-ornithine production by ornithine cyclodeaminase (Ocd) and that Ocd is a key enzyme for L-ornithine production under proline-supplemented conditions in C. glutamicum [12]. In our lab, we constructed a triple gene knockout strain C.glutamicum ATCC13032 ( ΔargFΔproBΔkgd) for L-ornithine production, which could produce 4.62 g/L of L-ornithine [13]. The level of L-ornithine production of the engineered strain is higher than that of the industrial strain Brevibacterium ketoglutamicum BK533 (2 g/L) under unoptimized conditions [14]. In order to further investigate the effect of conditions and to improve L-ornithine production, it is desirable to optimize the medium. It is well known that optimization of culture conditions is the primary task when developing industrial fermentation because they could strongly impact the product yield. Few papers reported the results of enhance of L-ornithine production. Lee et al. [15] reported that yeast extract as an arginine source and ammonium sulfate as an inorganic nitrogen source had significant effects on L-ornithine production and cell growth in Brevibacterium ketoglutamicum. Model of fed-batch culture, dilute rate and dual limitation of arginine and phosphate in continuum culture were also optimized for L-ornithine production and cell growth in Brevibacterium ketoglutamicum BK533 [7,14,16]. After these optimization, Brevibacterium ketoglutamicum BK533 production L-ornithine up to 74 g/L in a fed-batch culture by shorten the mixing time of the limiting nutrient in the fermenter [16] from 2 g/L in a batch culture [14].
However, to our knowledge, there is no report on optimization of L-ornithine production by engineering strain. Response surface methodology (RSM) has successfully been applied in optimization of bioprocess [17-22]. It can be used to evaluate the relationship between a set of controllable experimental factors and observed results. The interac- tion among the possible influencing parameters can be evaluated with limited number of experiments. In this study, statistical optimization of the fermentation medium was carried out in order to develop an industrial fermentation of L-ornithine by engineering Corynebacterium Glutamicum (ΔargFΔproBΔkgd). The preliminary one-factor-at-a-time approach was used to obtain the critical components of the medium. Then, the important factors were selected out by Plackett-Burman design. Finally, the RSM was employed to build models to evaluate the effective factors and to study their interaction and to select optimum conditions. L-ornithine production was enhanced by 197% under optimized conditions, compared to that under original conditions. On the other hand, the process of L-ornithine production of the engineered strain in a 5 L bioreactor was also characterized. L-ornithine production reached 18.17 g/L at 60 h of fermentation, which is the highest level in engineered strain at present.
Materials and Methods
Microorganism and medium
The engineering strain C.glutamicum ATCC13032 (ΔargFΔproB Δkgd) [13] was used in this study. The seed medium consisted of (per liter ) 25.00 g of glucose, 10.00 g of yeast extract, 10.00 g of corn steep liquor, 15.00 g of (NH4)SO4, 2.50 g of MgSO4·7H2O, 1.00 g of KH2PO4, 0.50 g of K2HPO4, 0.50 g of Na2HPO4 and 10.00 g of CaCO3. The initial fermentation medium consisted of (per liter) 100.00 g of glucose, 20.00 g of corn steep liquor, 50.00 g of (NH4)SO4, 2.50 g of MgSO4·7H2O, 1.00 g of KH2PO4, 0.50 g of K2HPO4, 0.50 g of Na2HPO4, 20 mg of FeSO4·7H2O, 20 mg of MnSO4·4H2O, 2.00 g of molasses, and 10.00 g of CaCO3. Initial PH of all the above media was adjusted to 7.0. All media were autoclaved for 20 min at 121°C. In this study, the composition of the fermentation medium varied according to the experimental design.
Cultivation
The seed medium was grown at 30°C in the rotary shaker at 150 rpm for 20 hours. For L-ornithine fermentation, a 1.0-ml sample of the seed culture was inoculated into 10 ml of the fermentation medium in a 100-ml rotary flask and cultivated at 30°C and 150 rpm for 72 hours. Triplicate experiments were carried out and the mean value was calculated.
Statistical optimization experimental design
Plackett-Burman design: Plackett-Burman design was two-level partial factorials most commonly employed for identifying important factors for further investigation. It based on the first order model:
Y = β0 + Σβixi (1)
This design does not consider the interaction effects among the variables. Fourteen variables were screened in sixteen experiments with one dummy variable. The experimental design is shown in Table 1 (Hadamard matrix). Each variable was represented in two levels namely a high level denoted by (+) and a low level designated by (−). All experiments were performed induplicate and the average of L-ornithine production after 72 h was taken as the response (Y). The variables whose confidence levels greater than 85 % were considered to significantly influence L-ornithine production.
| No. | Variables/levelsa | L-ornithine (g/L) |
||||||||||||||
| X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | X12 | X13 | X14 | X15 | ||
| 1 | + | + | + | + | + | + | + | + | - | - | - | - | - | - | - | 7.75 |
| 2 | - | + | + | + | + | + | + | + | + | - | - | - | - | - | - | 11.56 |
| 3 | - | - | + | + | + | + | + | + | + | + | - | - | - | - | - | 8.96 |
| 4 | - | - | - | + | + | + | + | + | + | + | + | - | - | - | - | 9.38 |
| 5 | - | - | - | - | + | + | + | + | + | + | + | + | - | - | - | 7.47 |
| 6 | - | - | - | - | - | + | + | + | + | + | + | + | + | - | - | 11.13 |
| 7 | - | - | - | - | - | - | + | + | + | + | + | + | + | + | - | 11.42 |
| 8 | - | - | - | - | - | - | - | + | + | + | + | + | + | + | + | 10.06 |
| 9 | + | - | - | - | - | - | - | - | + | + | + | + | + | + | + | 7.96 |
| 10 | + | + | - | - | - | - | - | - | - | + | + | + | + | + | + | 11.31 |
| 11 | + | + | + | - | - | - | - | - | - | - | + | + | + | + | + | 9.79 |
| 12 | + | + | + | + | - | - | - | - | - | - | - | + | + | + | + | 8.98 |
| 13 | + | + | + | + | + | - | - | - | - | - | - | - | + | + | + | 7.43 |
| 14 | + | + | + | + | + | + | - | - | - | - | - | - | - | + | + | 9.64 |
| 15 | + | + | + | + | + | + | + | - | - | - | - | - | - | - | + | 8.63 |
| 16 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 9.87 |
aX1, Glucose at a high level of 125 and a low level of 100; X2, K2HPO4 at a high level of 0.625 and a low level of 0.500; X3 , (NH4)2SO4 at a high level of 6.25 and a low
level of 5.00; X4, MgSO4∙7H2O at a high level of 3.125 and a low level of 2.500; X5, Yeast extract at a high level of 25 and a low level of 20; X6, KH2PO4 at a high level
of 1.25 and a low level of 1.00; X7, Na2HPO4 at a high level of 0.625 and a low level of 0.500; X8, FeSO4∙7H2O at a high level of 0.025 and a low level of 0.020; X9,
MnSO4∙4H2O at a high level of 0.025 and a low level of 0.020; X10, molasses at a high level of 2.5 and a low level of 2.0; X11, CaCO3 at a high level of 12.5 and a low
level of 10.0; X12, KAc at a high level of 6.25 and a low level of 5.00; X13, succinic acid at a high level of 6.25 and a low level of 5.00; X14, Tween80 at a high level of 1.25
and a low level of 1.00; X15, dummy variable.
Table 1: Plackett-Burman experiment design matrix for screening of culture of L-ornithine production
Steepest ascent path design: The design was begun from the center point of the Plackett-Burman design and increased as direction ratio of regression coefficients. The direction of steepest ascent was parallel to the response of Equation (1). The steepest ascent experiments were performed until no further increase of the response. This point was in the optimal range and was used as the center point for optimization using the central composite design.
Central composite design: The central composite designs are composite designs formed from two-level factorials by the addition of just enough points to estimate curvature and interaction effects. It was generated by Design Expert software (version 7.1.3, Stat-Ease Inc., Min neapolis, USA) to estimate the response of the dependent variable. In this study, A 23 factorial central composite experimental design with four start points (α = 1.68) and six replicates at the central point were adopted. According to this design, 20 experiments were used to optimize the screened variables grouped as yeast extract (x1), NH4 (SO4)2 (x2) and MgSO4·7H2O (x3). The experimental design is shown in Table 2 and Table 3. The variables were coded according to the equation (2):
| Factor | Code | Coding level | |||||
|---|---|---|---|---|---|---|---|
| Uncoded | Coded | -1.68 | -1 | 0 | 1 | 1.68 | |
| Yeast extract (g/L) | X1 | x1 | 12.64 | 14 | 16 | 18 | 19.36 |
| (NH4)2SO4 (g/L) | X2 | x2 | 31.6 | 35 | 40 | 45 | 48.4 |
| MgSO4×7H2O (g/L) | X3 | x3 | 1.76 | 1.9 | 2.1 | 2.3 | 2.44 |
Table 2: Range of different factors studies in the CCD design
| No. | x1 | x2 | x3 | L-ornithine concentration(g/L) | |
| Experiment value | Predicted value | ||||
| 1 | 1 | 1 | -1 | 11.99 | 12.13 |
| 2 | 0 | 0 | 0 | 14.02 | 13.87 |
| 3 | -1 | -1 | -1 | 14.07 | 14.04 |
| 4 | 0 | 0 | 0 | 13.94 | 13.87 |
| 5 | 0 | 0 | 1.68 | 13.84 | 13.91 |
| 6 | 0 | -1.68 | 0 | 13.67 | 13.75 |
| 7 | -1 | 1 | -1 | 12.60 | 12.61 |
| 8 | 0 | 0 | 0 | 13.96 | 13.87 |
| 9 | -1 | -1 | 1 | 14.02 | 13.89 |
| 10 | 0 | 0 | 0 | 13.79 | 13.87 |
| 11 | 0 | 0 | 0 | 13.90 | 13.87 |
| 12 | 0 | 1.68 | 0 | 12.644 | 12.57 |
| 13 | -1 | 1 | 1 | 13.56 | 13.54 |
| 14 | 1 | 1 | 1 | 13.41 | 13.44 |
| 15 | -1.68 | 0 | 0 | 13.65 | 13.77 |
| 16 | 0 | 0 | 0 | 13.59 | 13.87 |
| 17 | 0 | 0 | -1.68 | 13.01 | 12.93 |
| 18 | 1.68 | 0 | 0 | 13.07 | 12.96 |
| 19 | 1 | -1 | 1 | 13.40 | 13.41 |
| 20 | 1 | -1 | -1 | 13.16 | 13.18 |
Table 3: Central composite design matrix of independent variables with corresponding experimental and predicted value
xi = (Xi − X0)/?Xi = 1, 2, 3,..., j (2)
where xi = the coded value of the independent variable Xi, Xi =the uncoded value of the independent variable, X0 = the value of Xi at the center point, and ?X = the step change value.
The relationships of the variables were determined by fitting the second order polynomial equation to data obtained from 20 experiments using the mean values of the triplicates of each experiment conducted thrice at different occasions. The second order polynomial equation (3) was showed as follows:
y = β0 + Σβixi + Σβiixi2+ Σβijxixj (3)
where y = predicted response, β0 = offset term, βi = linear effect, βii = squared effect, and βij =interaction effect.
The Design Expert software was used for regression analysis of the data obtained and to estimate the coefficients of the regression equa tion. Iso-response contour plots were also obtained by using Design Expert software.
L-ornithine production of the engineered strain in the 5L bioreactor
For L-ornithine fermentation, 10% (v/v) of 20 h inoculum were inoculated into 3.5 L of the fermentation medium in a 5 L bioreactor (Biostat B5, B. Braun Biotech International Diessel GmbH, Germany) and cultivated at 30°C. NH4OH was used to control PH of the above medium. Antifoam was added manually as needed. The initial aeration rate was 1.0 L/L/min and the agitation rate was 800 rpm. When dissolved oxygen was below 15%, the aeration rate was increased to 1.5 L/L/min until the end of fermentation. OD600, residual glucose concentration and L-ornithine production assay of the broth were carried out every some time.
Analytical method
Cell growth was monitored by measuring the optical density of the culture at 600 nm (OD600) using a spectrophotometer (Shimadzu Corporation, Japan) after dilution of the culture with 0.2M HCl to dissolve CaCO3. L-ornithine concentrations were determined by the colorimetric method with Ninhydrin as described previously [13,23]. Glucose concentration was determined the colorimetric method based on the phenol-sulfuric acid reaction [24].
Results and Discussion
Effect of nitrogen source on L-ornithine production
The effects of different nitrogen sources on L-ornithine production in Corynebacterium Glutamicum ATTC13032 (ΔargFΔproBΔ kgd) were investigated using glucose as carbon source. The results of growth and L-ornithine production are shown in Table 4. For a single nitrogen source, the highest L-ornithine production was obtained with 2 % yeast extract. The complex nitrogen sources (yeast and ammonium sulfate) were more desirable than a single nitrogen source. The highest L-ornithine production (6.01 g/L) and growth (43.45 of OD600) were observed with 2 % yeast extract and 5% ammonium sulfate. Lee et al. reported that the highest L-ornithine production was obtained with 10 g/L yeast extract and 20 g/L ammonium sulfate in Brevibacterium ketoglutamicum [15].
| Nitrogen source | L-Ornithine (g/L)a | OD600a |
| 2 % Corn steep liquor | 3.25±0.01 | 17.38±0.58 |
| 2 % Yeast extract | 4.59±0.05 | 24.57±0.05 |
| 2 % Beef extract | 2.14±0.03 | 19.67±0.45 |
| 2 % Peptone | 2.24±0.03 | 25.76±0.06 |
| 2 % Urea | 1.13±0.01 | 6.37±0.10 |
| 5 % (NH4)2SO4 | 0.91±0.01 | 21.88±0.32 |
| 5 % NH4NO3 | 0.57±0.01 | 7.63±0.26 |
| 5 % NH4Cl | 0.80±0.01 | 13.78±0.20 |
| 2 %Yeast+5 % (NH4)2SO4 | 6.01±0.03 | 43.45±0.10 |
a Data represent the mean of the triplicate cultures±standard deviation
Table 4: Effects of various nitrogen sources on growth and L-ornithine production in Corynebacterium glutamicum ATTC13032 (ΔargFΔproBΔkgd)
Effect of tricarboxylic acid cycle intermediates on L-ornithine production
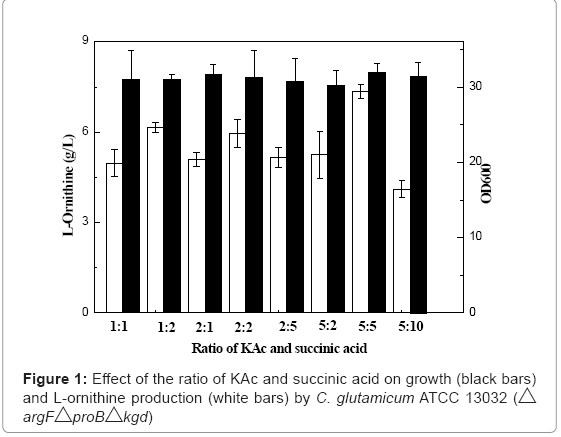
In C. glutamicum ATTC13032 (ΔargFΔproBΔkgd), the flux of the downstream of 2-oxoglutarate was weakened because of the deletion of kgd gene. Increasing the flux of the downstream of 2-oxoglutarate by supplement of tricarboxylic acid cycle intermediates may be beneficial to growth and production. Thus, we investigated the effect of tricarboxylic acid cycle intermediate (KAc and succinic acid) on L-ornithine production. Figure 1 shows that the effect of different ratio of KAc and succinic acid on growth and L-ornithine production. The highest Lornithine production and growth were obtained with 5:5 of the ratio of KAc and succinic acid. Addition of tricarboxylic acid cycle intermediates also resulted in the increase of production of poly (γ-glutamic acid) (36.9 %) by Bacillus licheniformis [25]. L-ornithine is formed from glutamate as poly (γ-glutamic acid). Wendisch et al. [26] showed a two- to fourfold higher carbon flux into tricarboxylic acid cycle when C. glutamicum cells were grown in the presence of acetate. Their further studies showed that the expression of genes of glyoxylate cycle (aceA and aceB) was upregulated in the presence of acetate [27]. Yu et al. [28] reported that glyoxylate cycle is required for the overproduction of glutamate and the glutamate production was insignificantly inhibited after aceA gene deletion.
Effect of trigger on L-ornithine production
Recent studies have demonstrated that a reduction in the 2-oxoglutarate dehydrogenase complex (ODHC) activity is important for glutamate production by C. glutamicum [29,30]. Moreover, only when triggering by Tween 40, the inactivation of ODHC could enhance glutamate production in C. glutamicum [30]. Thus, we investigated the effects of different trigger reagent (Tween 80, penicillin G, ampicilin, glycine and isonicotinic acid) on L-ornithine production in preliminary experiment and found that Tween 80 showed the best effect on L-ornithine production. Table 5 shows the effect of addition time of Tween 80 on growth and L-ornithine production. When Tween 80 was added into the medium after 8 h, the highest L-ornithine production (10.21 g/L) was obtained. Earlier addition inhibited growth and L-ornithine production and later addition had a negative effect only for L-ornithine.
| Time(h) | L-ornithine production (g/L)a | OD600a |
| 0 | 8.48±0.15 | 21.60±1.46 |
| 4 | 9.55±1.20 | 25.06±0.99 |
| 8 | 10.21±0.38 | 27.33±2.71 |
| 12 | 9.45±0.21 | 31.80±1.07 |
aData represent the mean of the triplicate cultures±standard deviation
Table 5: Effect of 0.1% Tween80 additive time on growth of C. glutamicum ATCC 13032 (ΔargFΔproBΔkgd) and L-ornithine production
Screening of culture conditions of the engineered strain for L-ornithine production by the Plackett-Burman Design
Based on the above results, fourteen variables, X1 (glucose), X2 (K2HPO4), X3 ((NH4)2SO4), X4 (MgSO4.7H2O), X5 (yeast extract), X6 (KH2PO4), X7 (Na2HPO4), X8 (FeSO4.7H2O), X9 (MnSO4· 4H2O), X10 (molasses), X11 (CaCO3), X12 (KAc), X13 (succinic acid), X14 (Tween 80) were chosen to be analyzed by the Plackett-Burman Design. The experimental design and corresponding L-ornithine production are shown in Table 1. When the sign of the effect Exi of the tested variable is positive, the influence of the variable on L-ornithine production is greater at a high level. And when negative, the effect of the variable is greater at a low level. As shown in Table 6, it was found that Na2HPO4, FeSO4·7H2O, MnSO4·4H2O, molasses, CaCO3, KAc, succinic acid and Tween 80 had a positive effect on L-ornithine production, while glucose, K2HPO4, (NH4)2SO4, MgSO4·7H2O, yeast extract and KH2PO4 showed a negative effect on L-ornithine production.
| Variable | Exi | txi | P |
| X1 | -8.37 | -2.23 | 0.04 |
| X2 | -1.19 | -0.32 | >0.50 |
| X3 | -5.88 | -1.57 | 0.14 |
| X4 | -6.69 | -1.79 | 0.10 |
| X5 | -9.72 | -2.59 | 0.02 |
| X6 | -2.31 | -0.62 | >0.50 |
| X7 | 1.26 | 0.34 | >0.50 |
| X8 | 4.11 | 1.10 | 0.31 |
| X9 | 4.53 | 1.21 | 0.26 |
| X10 | 4.06 | 1.08 | 0.32 |
| X11 | 5.71 | 1.52 | 0.16 |
| X12 | 4.90 | 1.31 | 0.21 |
| X13 | 4.83 | 1.29 | 0.22 |
| X14 | 1.84 | 0.49 | >0.50 |
Table 6: Results of the screening of culture of L-ornithine production
The variables with confidence levels greater than 85% were considered significant. glucose, (NH4)2SO4, MgSO4·7H2O and yeast extract were significant (>85 % confidence levels) for L-ornithine production, whereas Na2HPO4, FeSO4·7H2O, MnSO4·4H2O, molasses, CaCO3, KAc, succinic acid and Tween 80 were insignificant with positive effect on Lornithine production. Moreover, K2HPO4 and KH2PO4 were found insignificant with negative effect on L-ornithine production. These insignificant factors were not included in the next optimization experiment, but were used in all trials at their (−1) level for negative contribution and (+1) level for the positive contribution. Lee et al. [15] also reported the similar results, which yeast extract and (NH4)2SO4 are important factors for L-ornithine production in Brevibacterium ketoglutamicum.
Optimization by steepest ascent path
Coefficients of X1, X3, X4 and X5 were negative, meaning that the path of the steepest ascent should decrease the concentration respectively in order to enhance the L-ornithine production. The center point of the Plackett-Burman Design was considered as the origin of the path. The experimental design and responses of the steepest ascent path experiments were given in Table 7. It was clearly seen that the yield profile showed a maximum at run 3. Because glucose was used as carbon source and the skeleton of L-ornithine, (NH4)2SO4, MgSO4·7H2O and yeast extract were chosen for further optimization by RSM.
| Run | Yeast extract (g/L) | Glucose (g/L) |
(NH4)2SO4 (g/L) | MgSO4×7H2O (g/L) | L-ornithine concentration(g/L) |
| 1 | 20 | 100 | 50 | 2.5 | 10.59 |
| 2 | 18 | 90 | 45 | 2.3 | 11.24 |
| 3 | 16 | 80 | 40 | 2.1 | 13.33 |
| 4 | 14 | 70 | 35 | 1.9 | 13.08 |
| 5 | 12 | 60 | 30 | 1.7 | 12.98 |
| 6 | 10 | 50 | 25 | 1.5 | 9.13 |
| 7 | 8 | 40 | 20 | 1.3 | 9.52 |
Table 7: Steepest ascent experiment design and experimental result
Optimization of the screening culture conditions by response surface methodology
Yeast extract (x1), (NH4)2SO4 (x2), and MgSO4·7H2O (x3) were selected and optimized using central composition design (CCD). The CCD of three variables in coded and real levels was shown in Table 2. The experimental design and the experimental and predicted response of the L-ornithine production at 3 days were shown in Table 3. By applying multiple regression analysis on the data obtained from the CCD experiment, the following second order polynomial equation was proposed to calculate the optimum levels of these variables and to explain the L-ornithine production.
y =13.87-0.24 x1-0.35 x2+0.29 x3+0.095 x1 x2+0.094 x1 x3+0.27x2 x3- 0.18x12-0.25 x22-0.16 x32 (4)
Statistical testing of the model was done by the Fisher’s statistical test for analysis of variance (ANOVA) and the results were presented in Table 8. The fit of the model was checked by the coefficient of determination (R2), which was calculated to be 96.62%, indicating that 96.62 % of the total variability in the response could be explained by this model. Values of ‘Prob > F’ less than 0.05 indicate model terms are significant.
| Source | Sum of Squares | df | Mean Square | F Value | Prob > F |
|---|---|---|---|---|---|
| Model | 5.85 | 9 | 0.65 | 31.72 | < 0.001 |
| Pure Error | 0.12 | 5 | 0.024 | ||
| Corrected total | 6.06 | 19 |
aCoefficient of variation (CV) = 1.06 %; coefficient of determination (R2) =96.62%; correlation coefficient (R) = 98.30%, and adjusted R2=93.57%.
Table 8: Analysis of variance (ANOVA) for the selected quadratic modela
The Student t-distribution and the corresponding P-values, along with the parameter estimate, are given in Table 9. The P-value is the probability of obtaining a test statistic at least as extreme as the one that was actually observed, assuming that the null hypothesis is true. The lower the P-value, the less likely the result is, and consequently the more significant the result is, in the sense of statistical significance. The P-values are necessary to understand the pattern of the mutual interactions between the best variables. The parameter estimate and the corresponding P-values (Table 9) suggest that all the independent variables including yeast extract (x1), (NH4)2SO4 (x2), and MgSO4·7H2O (x3) had a significant effect on the L-ornithine production. The quadratic term of all variables including x12, x22, x3 2 also were significant and the mutual interaction terms of x2x2 had a significant effect on L-ornithine production. The results indicate that L-ornithine production was increased with the decrease of yeast extract concentration, and that L-ornithine production was increased with the decrease of (NH4)2SO4 concentration and then decreased when (NH4)2SO4 concentration dropped below some value. It also indicates that L-ornithine production was increased with the increase of MgSO4·7H2O concentration and then decreased when MgSO4·7H2O concentration increased above some value.
| Model term | Parameter estimate | Degree of freedom | Computed t value | P value |
| β0 | 13.87 (5.8) | 1 | ||
| x1 | -0.24 (3.9) | 1 | -0.062 | <0.001 |
| x2 | -0.35 (3.9) | 1 | -0.090 | <0.001 |
| x3 | 0.29 (3.9) | 1 | 0.074 | <0.001 |
| x1* x2 | 0.095 (5.1) | 1 | 0.019 | 0.089 |
| x1* x3 | 0.094 (5.1) | 1 | 0.018 | 0.094 |
| x2* x3 | 0.27 (5.1) | 1 | 0.053 | <0.001 |
| x12 | -0.18 (3.8) | 1 | -0.047 | 0.001 |
| x22 | 0.25 (3.8) | 1 | 0.066 | <0.001 |
| x32 | -0.16 (3.8) | 1 | -0.042 | 0.002 |
Table 9: The least-squares fit and the parameter estimates (significance of regression coefficient)
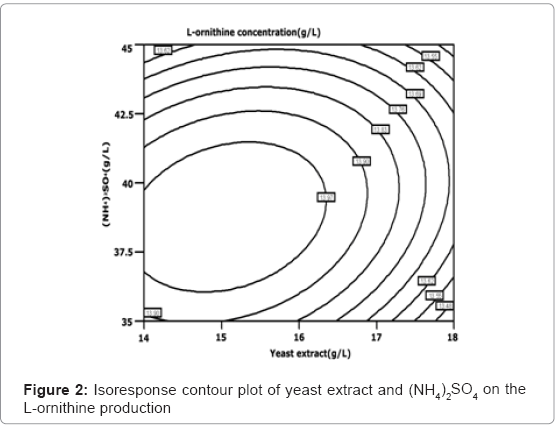
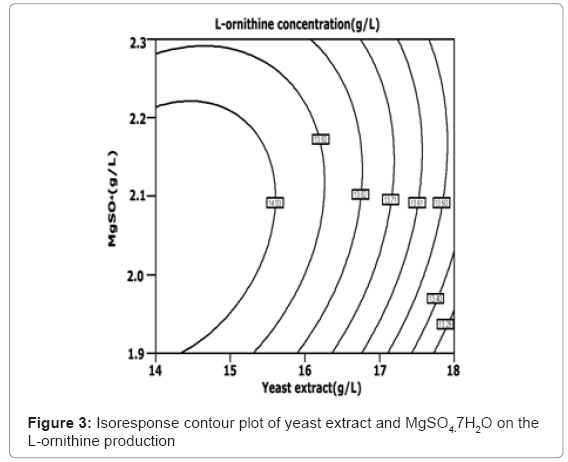
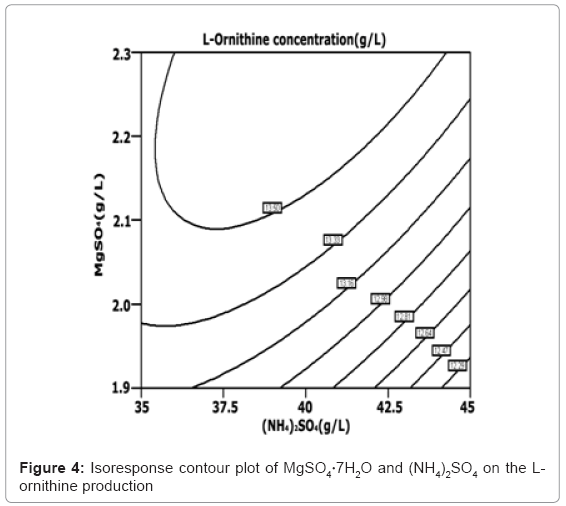
The isoresponse contour plot is shown in Figure 2, Figure 3 and Figure 4, respectively. Figure 2 shows the relative effect of yeast extract and (NH4)2SO4 on the L-ornithine production. Figure 3 shows the relative effect of yeast extract and MgSO4·7H2O on the L-ornithine production. The less spherical isoresponse contour plot in Figure 3 and 4 also indicates that the mutual interaction terms of x1x2 and x1x3 were insignifi-cant. Figure 4 shows the relative effect of MgSO4·7H2O and (NH4)2SO4 on the L-ornithine production, indicating the mutual interaction between MgSO4·7H2O and (NH4)2SO4 was significant. The regression equation (Equation (4)) is solved by using Design Expert software. The optimal concentration of yeast extract, (NH4)2SO4 and MgSO4·7H2O is 13.99, 37.94, and 1.58 g/L, respectively. The maximum predicted value of the L-ornithine yield obtained is 14.13 g/L.
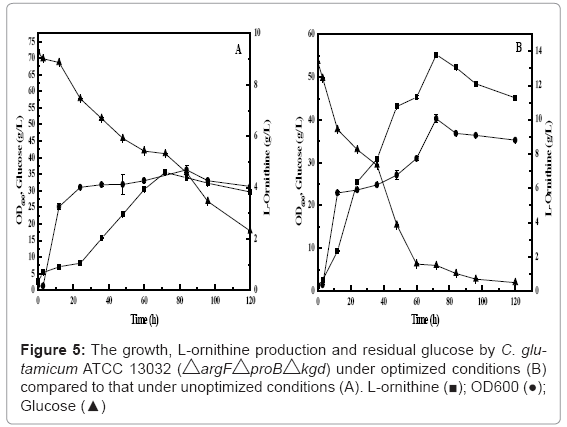
Comparison of the L-ornithine fermentation between under optimal conditions and original conditions was carried out. The results of the cell growth, L- ornithine production and residual glucose by the engineered strain were given in Figure 5. The maximal L-ornithine level obtained was 13.73 g/L. This value was close to the predicted value. The optimization resulted in 197% increase of the L-ornithine production (from 4.62 g/L under unoptimized conditions to 13.73 g/L under optimized conditions). Optimization also resulted in the increase of the cell growth (from 37.75 of OD600 under unoptimized conditions to 40.25 of OD600 under optimized conditions). Cell growth was very fast in the first day and then became slow to the end of fermentation. Optimization also resulted in the decrease of the glucose consumption rate (from 0.045 g/L h under unoptimized conditions to 0.039 g/L h under optimized conditions). And the glucose concentration dropped below 10.00 g/L under optimized conditions after 60 h.
L-ornithine production of the engineered strain in the 5 L bioreactor
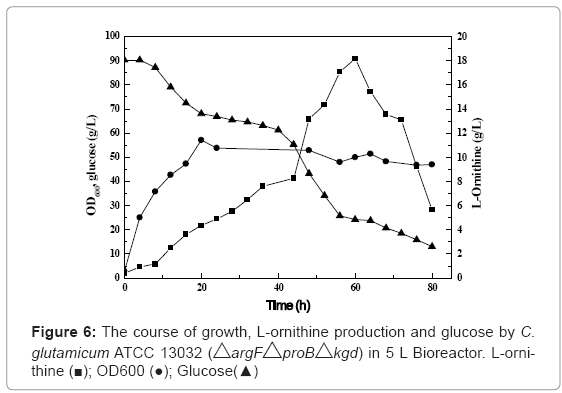
The results of the cell growth, L-ornithine production and residual glucose by the engineered strain in the 5 L bioreactor are presented in Figure 6. Cell growth rapidly went into exponential growth phase and then into station phase after 20 h. The glucose consumption rate increased significantly after 3 h, and then reduced between 20 and 40 h. Thereafter, it increased again in order to produce L-ornithine and finally decreased after 56 h. The level of L-ornithine increased progressively at the first 2 days and then enhanced sharply to reach the maximum 18.17 g/L at 60 h. From Figure 6, we can also find that the concentration of L-ornithine became to decrease sharply after 60 h, whereas glucose was still consumed. We also found that the value of PH after 60 h also increased rapidly (Data not shown). The reason may be that L-ornithine could be converted to polyaminime because the expression level of spermidine synthase of the engineered strain cultured in flask after 72 h was about 5 fold higher than that of the parent strain ATCC 13032 [13]. But the real reason should be further study.
The maximum L-ornithine production of 18.17 g/L at 60 h, which is unprecedented in engineered strain, was obtained. The level of Lornithine production is 9 times higher than that (2 g/L) of the classically obtained auxotrophic mutant Brevibacterium ketoglutamicum BK533 in a batch culture [14]. However, it is lower than that (74 g/L) of the auxotrophic mutant Brevibacterium ketoglutamicum BK533 in a fed-batch culture by shorten the mixing time of the limiting nutrient in the fermenter [16]. The engineered strain should be further modified by knockout and overexpression of key genes to improve L-ornithine production. The yield may be also enhanced by fed-batch fermentation technology. The further modification of the engineered strain and fedbatch fermentation are currently underway in our lab. The results will be reported shortly.
Conclusions
Optimization of fermentation conditions was carried out by onefactor- at-a-time, Plackett-Burman, steepest ascent design and response surface methodology. Tween 80, tricarboxylic acid cycle intermediates (KAc and succinic acid) were beneficial to L-ornithine production. The important factors, glucose, (NH4)2SO4, MgSO4·7H2O and yeast extract, were selected out using Plackett-Burman design. Then their response region close to the maximum production was obtained by using steepest ascent design. An optimized fermentation medium was obtained by using response surface methodology (g/L): 37.94 (NH4)2SO4, 1.58 MgSO4·7H2O and 13.99 yeast extract. The optimization resulted in the increase of L-ornithine production of 197 % compared with that under original conditions. 18.17 g/L of L-ornithine production was obtained in the 5-L bioreactor at 60 h, which is the highest level in engineered strain at present.
Acknowledgements
We are grateful to the Natural Science Foundation of China (Grant No. 30970089, 20876181, 20831006) and Natural Science Foundation of Guangdong Province (No. 9351027501000003) for their financial support.
References
- Hayashi M, Ohnishi J, Mitsuhashi S, Yonetani Y, Hashimoto S, et al. (2006) Transcriptome analysis reveals global expression changes in an industrial Llysine producer of Corynebacterium Glutamicum. Biosci Biotechnol Biochem 70: 546-550.
- Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104: 155-172.
- Park SD, Lee JY, Sim SY, Kim Y, Lee HS (2007) Characteristics of methionine production by an engineered Corynebacterium Glutamicum strain. Metab Eng 9: 327-336.
- Salvatore F, Cimino F, Maria C, Cittadini D (1964) Mechanism of the protection by L-ornithine-L-aspartate mixture and by L-arginine in ammonia intoxication. Arch Biochem Biophys 107: 499-503.
- Shi HP, Fishel RS, Efron DT, Williams JZ, Fishel MH, et al. (2002) Effect of Supplemental ornithine on wound healing. J Surg Res 106: 299-302.
- Zajac A, Poprzecki S, Zebrowska A, Chalimoniuk M, Langfort J (2010) Arginine and ornithine supplementation increases growth hormone and insulin-like growth factor-1 serum levels after heavy-resistance exercise in strength-trained athletes. J Strength Cond Res 24: 1082-1090.
- Choi DK, Ryu WS, Choi CY, Park YH (1996) Production of L-ornithine by arginine auxotrophic mutants of Brevibacterium ketoglutamicum in dual substrate limited continuous culture. J Ferment Bioeng 81: 216-219.
- Kinoshita S, Nakayama K, Udaka S (1957) The fermentative production of Lornithine. J Gen Appl Microbiol 3: 276-277.
- Zhang JF, Wang JB, Huang JM, Zhang J (2009) Breeding of high-yield L-ornithine- producing strain by protoplast fusion. Amino acids Biotic Resour 31: 53-57.
- Lee YJ, Cho JY (2006) Genetic manipulation of a primary metabolic pathway for L-ornithine production in Escherichia coli. Biotechnol Lett 28: 1849-1856.
- Hwang JH, Hwang GH, Cho JY (2008) Effect of increased glutamate availability on L-ornithine production in Corynebacterium Glutamicum. J Microbiol Biotechnol 18: 704-710.
- Lee SY, Cho JY, Lee HJ, Kim YH, Min J (2010) Enhancement of ornithine production in proline-supplemented Corynebacterium Glutamicum by ornithine cyclodeaminase. J Microbiol Biotechnol 20: 127-131.
- Lu DM, Liu JZ, Mao ZW Engineering of Corynebacterium Glutamicum to Enhance L-Ornithine Production by Gene Knockout and Comparative Proteomic Analysis. Chinese J Chem Eng, In press
- Choi DK, Ryu WS, Chung BH, Hwang SO, Park YH (1995) Effect of dilution rate in continuous production of L-ornithine by an arginine auxotrophic mutant. J Ferment Bioeng 80: 97-100.
- Lee TH, Chang YK, Ryu WS, Chung BH, Park YH (1996) Effects of medium components on L-ornithine production by Brevibacterium ketoglutamicum. Biotechnol. Bioprocess Eng 1: 41-45.
- Lee HW, Yoon S, Jang H, Kim C, Kim T, et al. (2000) Effects of mixing on fedbatch fermentation of L-ornithine. J Biosci Bioeng 89: 539-544.
- Xiong YH, Liu JZ, Song HY, Ji LN (2004) Enhanced production of extracellular ribonuclease from Aspergillus niger by optimization of culture conditions using response surface methodology. Biochem Eng J 21: 27-32.
- Liu JZ, Weng LP, Zhang QL, Xu H, Ji LN (2003) Optimization of glucose oxidase production by Aspergillus niger in a benchtop bioreactor using response surface methodology. World J Microbiol Biotechnol 19: 317-323.
- Roukas T, Niavi P, Kotzekidou P (2011) A new medium for spore production of Blakeslea trispora using response surface methodology. World J Microbiol Biotechnol 27: 307-317.
- Azma M, Mohamed MS, Mohamad R, Rahim RA, Ariff AB (2011) Improvement of medium composition for heterotrophic cultivation of green microalgae, Tetraselmis suecica, using response surface methodology. Biochem Eng J 53: 187-195.
- Singh AK, Chhatpar HS (2010) Optimization of protease production by Streptomyces sp. A6 using statistical approach for reclamation of shellfish waste. World J Microbiol Biotechnol 26: 1631-1639.
- Wang H, Jiang P, LuY, Ruan Z, Jiang R, et al (2009) Optimization of culture conditions for violacein production by a new strain of Duganella sp. B2. Biochem Eng J 44: 119-124.
- Chinard FP (1952) Photometric estimation of proline and ornithine. J Biol Chem 199: 91-95.
- Dubois M, Gilles KA, Hamilton JK, Reberse PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28: 350-356.
- Bajaj IB, Singhal RS (2009) Enhanced production of poly (γ-glutamic acid) from Bacillus licheniformis NCIM 2324 by using metabolic precursors. Appl Biochem Biotechnol 159: 133-141.
- Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ (2000) Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium Glutamicum during growth on acetate and/ or growth. J Bacteriol 182: 3088-3096.
- Gerstmeir R, Wndisch VF, Schnicke S, Ruan H, Farwick M, et al. (2003) Acetate metabolism and its regulation in Corynebacterium Glutamicum. J Biotechnol 104: 99-122.
- Yu BQ, Shen W, Wang ZX, Zhuge J (2005) Glyoxylate cycle is required for the overproduction of glutamate but is not essential for Corynebacterium Glutamicum growth on glucose. Chinese J Biotechnol 21: 270-274.
- Asakura Y, Kimura E, Usuda Y, Kawahara Y, Matsui K, et al. (2007) Altered metabolic flux due to deletion of odhA causes L-glutamate overproduction in Corynebacterium Glutamicum. Appl Environ Microbiol 73: 1308-1319.
- Kim J, Hirasawa T, Sato Y, Nagahisa K, Furusawa C, et al. (2009) Effect of odhA overexpression and odhA antisense RNA expression on Tween-40-triggered glutamate production by Corynebacterium Glutamicum. Appl Microbiol Biotechnol 81: 1097-1106.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 16817
- [From(publication date):
December-2011 - Nov 25, 2025] - Breakdown by view type
- HTML page views : 11928
- PDF downloads : 4889