Research Article Open Access
Operational Evaluation of the Rapid Viability PCR Method for Post-Decontamination Clearance Sampling
Staci Kane1, Sanjiv Shah2*,#, Sonia Létant1, Gloria Murphy1, Teneile Alfaro1, Julie Avila1, Edmund Salazar1, Marissa Mullins3 and Tonya Nichols2
1Lawrence Livermore National Laboratory, Livermore, CA, USA
2US Environmental Protection Agency, National Homeland Security Research Center, USA
3US Environmental Protection Agency, Office of Emergency Management USA
- *Corresponding Author:
- Sanjiv Shah
US Environmental Protection Agency
National Homeland Security Research Center
EPA-NHSRC, 1200 Pennsylvania Ave.
NW 8801 RR, Washington, DC 20460, USA
Tel: 202-564-9522
Fax: 202-564-1614
E-mail: shah.sanjiv@epa.gov
Received Date: April 16, 2013; Accepted Date: June 03, 2013; Published Date: June 06, 2013
Citation: Kane S, Shah S, Létant S, Murphy G, Alfaro T, et al. (2013) Operational Evaluation of the Rapid Viability PCR Method for Post-Decontamination Clearance Sampling. J Bioterr Biodef S3:016. doi:10.4172/2157-2526.S3-016
Copyright: © 2013 Kane S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioterrorism & Biodefense
Abstract
The Rapid Viability Polymerase Chain Reaction (RV-PCR) method was evaluated during the Bio-Response Operational Testing and Evaluation (BOTE), an interagency project to evaluate field-level facility biological remediation, using leading decontamination technologies. The tests were performed using an intentional release (aerosolization) of spores of Bacillus atrophaeus subspecies globigii (BG), as a surrogate for Bacillus anthracis, the etiologic agent for anthrax. Three decontamination methods were assessed including fumigation with vaporized hydrogen peroxide (VHP), fumigation with chlorine dioxide (CD), and a surface treatment process using pH-adjusted bleach.The RV-PCR method was developed to rapidly detect live B. anthracis spores during a bioterrorism event. The method uses a change in realtime PCR response before and after a nine hour incubation step, to determine the presence of viable bacterial spores in the sample; the method was recently verified for air filter, wipe and water samples at the 10-spore level for B. anthracis Ames spores, and was also developed for swab, sponge-stick, and vacuum sock/filter samples. In the method, high throughput sample processing is combined with PCR-based analysis before and after a rapid culture step to speed viability determination, especially for complex surface and environmental samples that present challenges to current culture-based methods. In the BOTE project, a total of 159 surface wipe samples from post-decontamination events were analyzed by splitting the suspension after spore recovery into two equal parts, with one part analyzed by RV-PCR and the other part by culture after concentrating to the same volume. In the BOTE project, the RV-PCR method provided rapid results for post-decontamination samples that were 98% (156/159 samples) consistent with results from culture analysis. The percentage agreement was noteworthy, given the large number of samples containing low spore levels. For the Post-VHP, Post-Bleach, and Post-CD event samples, the percentage agreement was 93% (41/44 samples), 100% (47/47 samples), and 100% (68/68 samples), respectively. The RV-PCR method performed well for the surrogate BG spores exposed to decontaminants at real-world application levels, and with wipe samples containing background debris and indigenous microbial populations.
Keywords
Bacillus anthracis; Anthrax; Bacterial spores; Viability; Vaporous hydrogen peroxide; Chlorine dioxide; pH-adjusted bleach; Real-time PCR
Introduction
To protect human health and ensure that the environment and facilities are restored to ambient conditions following a biothreat agent release, rapid viability testing methods are needed. This critical need was highlighted during the response to the 2001 anthrax attacks, in which clearance sampling and analysis represented a significant bottleneck, leading to delays to safely reopen facilities. In the event of a biothreat agent release, hundreds to thousands of samples of diverse types (surface and environmental) would need to be rapidly processed, in order to both characterize the extent of contamination and determine the efficacy of decontamination activities. Rapid results from post-decontamination sampling are also needed to determine whether decontamination was complete or incomplete, and therefore would require remobilization of disinfection equipment followed by additional sampling and analysis for clearance. Current viability methods are not only low throughput, but also are too labor-, space-, and time-intensive to meet the need for rapid analysis; current culturebased protocols require 18-24 hr for colony development, followed by confirmation via real-time PCR or serological methods [1,2]. Rapid Viability (RV)-PCR combines high throughput sample processing with real-time PCR analysis of the sample at the initial spore recovery, and following a rapid incubation step (9 hr), using the change in PCR response between time points to determine whether viable spores were present. The RV-PCR method presents an additional preparedness to meet the challenge for analysis of a large number of samples.
Manual and semi-automated RV-PCR protocols were initially developed and tested with surrogate organisms, including Bacillus atrophaeus (B. globigii, BG) and the attenuated B. anthracis Sterne strain for a range of sample types. Method development with BG spores included limited testing with chlorine dioxide fumigation, which showed good agreement between RV-PCR and culture results even for several partial-kill disinfectant levels [3]. In addition, a most-probablenumber variation of the method (MPN RV-PCR) was used in parallel with traditional culture analysis to provide a quantitative estimate of B. anthracis Sterne spore levels from macrofoam swab samples [4]. Results from the MPN RV-PCR method (average MPN values) were within an order of magnitude of the CFU values measured by the culture method, which was validated in a multi-laboratory study [1]. An integrated culture and real-time PCR method to assess viability of disinfectant-treated BG spores, using robotics and the MPN approach was also tested by Varughese et al. [5]. To improve the RV-PCR method, with regard to US EPA’s need for rapid analysis, the endpoint for the method was shortened from 16 hours to 9 hours by incorporating DNA extraction and purification steps, thus yielding viability results from up to 24 samples and controls in <15 hr [6]. The optimized method was developed and evaluated for virulent B. anthracis (Ames) spores, showing a limit of detection (LOD) at the 10-spore level for air filter, wipe, and water samples using manual and semi-automated protocols, even in the presence of high loadings of debris, high populations of dead target spores, and high populations of live, non-target cells and spores.
Since RV-PCR includes a culture or enrichment step, the approach is more consistent with current culture-based methods, and therefore, may be more readily accepted by decision-makers for post-decontamination clearance. Further, the approach is built on fundamental principles in microbiology (spore germination and outgrowth in broth culture) and molecular biology (measurement of target genomic DNA by real-time PCR), but needs to be validated for operational use with diverse sample types and backgrounds, containing microbiological, chemical, and physical challenges, including those from post-decontamination field samples. Other viability approaches using photo-reactive DNA intercalating dyes such as propidium monoazide (PMA) that preferentially enter dead cells [7], or spores [8], to block subsequent PCR amplification have shown promise for rapid analysis; however, test conditions (e.g. dye amount, exposure time, and photo-activation conditions) must be optimized and standardized for different applications (target organisms and sample matrices), and the method LOD may not be sufficient for clearance. In addition, PMA has been shown to enter live cells to some extent [9], therefore, potentially leading to underestimation of the viable target population. Other methods that detect dipicolinic acid (DPA) released during early spore germination, using measurement of Tb3+-DPA photoluminescence [10], or that measure ATPase activity from germinated spores [11] provide rapid viability analysis, but are not specific to B. anthracis spores.
Since the intended application for the RV-PCR method is for the analysis of samples collected after decontamination of a bioterrorism event site, it was important to confirm the method’s accuracy under actual decontamination scenarios. The Bio-Response Operational Testing and Evaluation (BOTE) field-level decontamination assessment afforded a unique opportunity to evaluate the RV-PCR method in comparison with the culture method for post - decontamination samples, and also to guide further research for method optimization. The BOTE Phase I project was a multi-agency effort designed to operationally test and evaluate three leading bacterial spore decontamination technologies, namely surface disinfection with pH-adjusted bleach and fumigation with either vaporous hydrogen peroxide (VHP) or chlorine dioxide (CD), in a full-scale exercise.
This paper presents data on the performance of the RV-PCR method during the BOTE. It should be noted that each wipe sample was split into two equal parts after sample processing and spore recovery: one part was analyzed by RV-PCR and the other by the traditional culture method upon concentration to the same volume. While this procedure might have compromised the results for the low spore number samples, it was necessary to enable the most appropriate method performance comparison. Therefore, any conclusions on the performance of the RVPCR research method during this evaluation must be carefully derived. Also, it should be noted that the current version of the RV-PCR method provides qualitative analyses (detection/non-detection of live spores) of the samples. Furthermore, the goal of this study was to compare RVPCR with traditional culture side-by-side with post-decontamination surface samples, and not to determine disinfectant efficacy.
Materials and Methods
Evaluation of extraction buffers used for spore recovery and culture analysis
Prior to sample analysis for the BOTE project, preliminary experiments were conducted to confirm the buffers to be used in the combined culture/RV-PCR protocol. Since the RV-PCR extraction buffer contains 30% ethanol (with 70% of 0.25 mM KH2PO4/0.1% Tween 80 [6]), an experiment was conducted to compare plate counts for sample extraction buffers, with and without ethanol, to confirm that ethanol did not negatively affect the culture results. In this experiment, 104 B. atrophaeus (ATCC #9372; Apex Laboratories, Apex, NC) spores were added to extraction buffer with or without 30% ethanol, serial dilutions were performed in the appropriate buffer, and 100 µL were spread onto six replicate Tryptic Soy Agar (TSA) plates for each buffer type. Plates were incubated overnight at 35°C, with colony-forming units (CFU) obtained the next day and corrected for dilution.
Since preliminary experiments showed significant foaming upon vortex mixing with extraction buffer containing 70% of 0.25 mM KH2PO4/0.1% Tween-80 [6], tests were performed to determine whether a modified extraction buffer with the Tween-80 concentration reduced to 0.05% negatively affected BG spore recovery. In addition, a second spore extraction step using 30% ethanol/70% 0.25 mM KH2PO4 was added, in order to improve the recovery efficiency and to yield sufficient sample volumes for parallel RV-PCR and culture analyses. The second spore extraction lacked Tween since sufficient Tween remained in the sample and associated liquid to keep spores dispersed for accurate pipetting. The change in buffers reduced the foaming of the recovered spore suspension, thus enabling both analyses to take place with sufficient volume.
To confirm the buffer modification did not negatively influence spore recovery, the standard buffer (30% ethanol/70% of 0.25 mM KH2PO4/0.1% Tween 80) used for both first and second spore extraction steps was analyzed in parallel with the modified buffers, as described above. For the experiment, eight replicate wipe samples containing 250 mg autoclaved AZ Fine Test Dust (Powder Technology, Inc., Burnsville, MN) spiked with 105 spores per sample were used for each buffer treatment. B. atrophaeus spores from Apex Laboratories were used, since the actual test BG spores were received later. No differences were expected between spore preparations in phosphate-buffered saline with Tween (PBST; Teknova, Cat. No. P0201), with regard to buffer effects on spore recovery. After performing the spore extraction steps with the appropriate buffers, serial dilutions were performed and three replicates of 100 µL each were spread onto TSA plates and incubated at 35ºC overnight; plate counts were obtained the next day, corrected for dilution and average CFU per sample were compared.
Analysis of the BG spore preparation used for dissemination
BG spores were prepared by the Critical Reagents Program (CRP) of the Joint Program Executive Office of the Department of Defense for use in the BOTE project. Since the CRP BG spore preparation contained high levels of exogenous DNA, the spores were first tested directly by real-time PCR analysis to determine the DNA background level, either associated with the spore coat or present in the formulation that could impact the RV-PCR method. In addition, the CRP BG spores were spiked onto wipe samples, to determine if the background DNA affected the PCR response at the initial time point (T0), and if any issues with method accuracy were observed at the method endpoint after nine hours incubation (T9).
For the testing, the US EPA supplied a ~ 0.1 g sample (104 mg) of the CRP BG spore preparation. This aliquot was then suspended in 1 mL PBST, serial dilutions were performed in PBST, and CFU per mass of dry material were determined by spread plating onto TSA (with incubation at 35°C). Colony counts showed that the original spore stock (~ 104 mg/mL) contained approximately 3.55×1010 CFU/ mL (referred to as the 10-1 dilution), or approximately 3.41×1011 CFU/g. Appropriate dilutions were prepared and spore levels from 101-107 were added to Tryptic Soy Broth (TSB) medium for direct PCR analysis, or added to wipe samples for RV-PCR analysis. For direct analysis of spore suspensions, 5 µL of the spore dilution (in PBST) was added directly to 20 µL of PCR mix and analyzed using the BG real-time PCR assay (see details below). For RV-PCR analysis with BG dilutions spiked onto wipe samples, the protocol as outlined below was followed, with the exception that 13 mL were removed for each spore extraction step, instead of 14 mL. For real-time PCR analysis, B. atrophaeus genomic DNA standards were included to estimate genome equivalents present in the spore preparation. This testing also evaluated the algorithm for determining presence/absence of viable spores, which included the Ct at T9 and the ΔCt (i.e. Ct[T0]Ct[T9])-to determine if any adjustments were needed for the BOTE project.
Sample type
Wipe samples used in the study were 2”×2” (5.1×5.1 cm) wipes (Kendall™, Versalon™, Cat. No. 8042, 50% rayon and 50% polyester gauze). Pre-wetted, sterile wipes were prepared at LLNL and provided to EPA field sampling personnel. Briefly, a sterile gauze wipe was placed into a 30 mL tube with screw cap (E&K Scientific, Cat. No. EK-T324S), and 1.5 mL of sterile PBST buffer were added to the wipe. Each tube was bar-coded, placed into a 4” ×6” zip lock bag, and a replicate barcode was placed on the bag. In addition, pre-wetted wipe samples remained at LLNL for use as True Blanks (TB). Prior to shipment to field personnel, random wipe samples were tested for sterility by washing in buffer and spread plating onto TSA. No growth was observed after incubation for two days at 35°C.
Spore dissemination, decontamination, and sampling
The BOTE project was conducted in a two-story unoccupied office building with rooms containing both porous and nonporous surfaces, simulating commercial office and residential environments. In the areas where sampling for RV-PCR analysis was conducted, spores were disseminated at approximately 1×106 CFU per ft2 using wet aerosol dispersion (nebulization). After overnight settling, pre-decontamination sampling was conducted, followed by decontamination, post-decontamination sampling, and re-setting the facility for testing the next decontamination technology. For this study, fresh gloves were donned and pre-wetted wipes were removed from the 30 mL tubes and used to sample a 1 ft2 area following EPA sampling protocols.
Field and laboratory controls
Field quality control samples, referred to as QC samples, were opened during sampling, but were not used to wipe surfaces. These were included with batches of surface samples during shipment, and were not marked as QC samples. Laboratory controls included “True Blanks”, positive controls (PCs), and negative controls (NCs). The PCs consisted of wipes spiked with BG spore suspension (Apex Laboratories, Inc.; presently Yakibou Laboratories) in PBST buffer; approximately 105 CFU per PC were used for VHP test sample batches, which was later adjusted to 50-100 CFU per PC for bleach and CD test sample batches. The NCs were pre-wetted wipes spiked with the same volume of PBST buffer only.
Sample receiving
Sample shipments by overnight cold-pack were coordinated by staff at the Idaho National Laboratory (INL). The chain-of-custody (COC) forms provided with each shipment did not state whether wipes were from surface sampling or whether they were QC samples, in order to maintain the scientific integrity of the test. During receiving, outer surfaces of bags containing sample tubes were bleached, and the tube was bleached, as it was removed from the bag and placed into a tube rack. The barcode was scanned according to the position in the rack. One sample was processed at a time and gloves were changed between samples to prevent cross-contamination. Prior to conducting spore recovery, a sterile forceps was used to place the mesh support over the wipe sample, to keep the wipe clear of pipetting activities. Previous work had shown that extraction efficiency could also be enhanced when a mesh support was used to hold the wipe to the side of the tube, allowing buffer to be washed through the wipe sample during vortexing [3].
Wipe sample processing
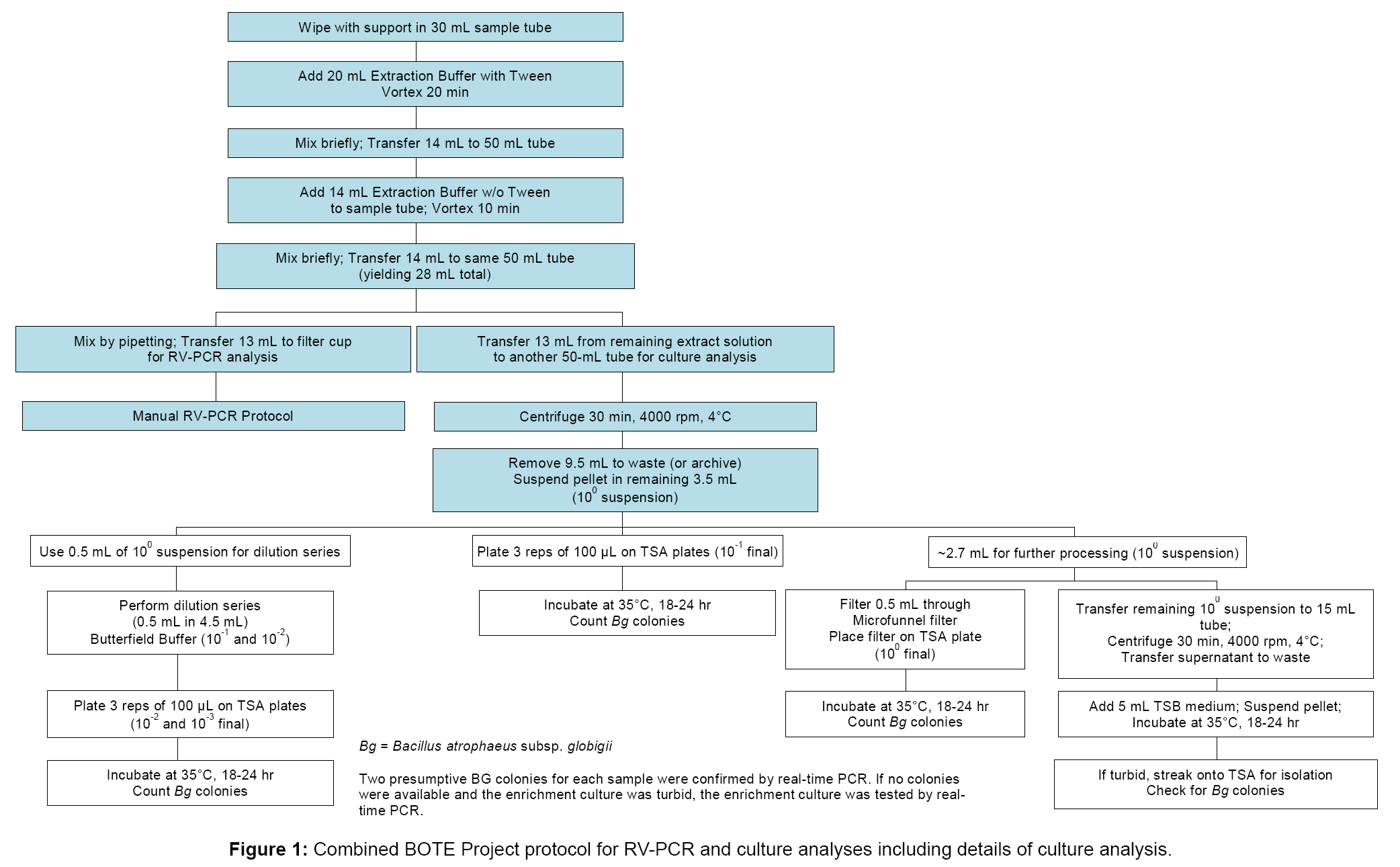
The single-laboratory verified RV-PCR protocol for detection of viable B. anthracis from wipe samples served as a basis for the BOTE RV-PCR method evaluation [6]. In addition, modifications were made to allow culture analysis in parallel with RV-PCR analysis, as outlined in Figure 1, with detailed steps for RV-PCR analysis shown in Figure 2. To accommodate both analyses, the sample extract following spore removal was split into two aliquots, with one aliquot used for RV-PCR analysis, and the other aliquot used for culture analysis. The manual RV-PCR protocol was used, since samples were received and processed in smaller batches (up to 48 per batch), and use of the previously developed semi-automated RV-PCR protocol was not warranted. For RV-PCR analysis, the optimized protocol for B. anthracis detection, using a nine hour incubation and DNA extraction and purification (modified Promega® MagneSil® method using paramagnetic particles, PMP), was adapted for BG detection. The aliquot for culture analysis was concentrated by centrifugation to the same extent as that for RVPCR analysis, followed by serial dilution and plating, microfunnel filtration, and enrichment of the remaining suspension.
RV-PCR sample processing and analysis
For RV-PCR analysis which uses sample tube racks accommodating 24 tubes, up to 21 samples were processed per rack along with one NC, one PC, and at least one TB. One to three TBs were included for each set of samples. To each wipe sample in a 30 mL tube, 20 mL of cold Extraction Buffer with Tween (70% of 0.25 mM KH2PO4/0.05% Tween 80 (pH 7.4) and 30% ethanol; final pH ~ 9.5) were added, and the tubes were vortexed for 20 min on a platform vortexer, set to speed 7 (e.g. VWR Cat. No. 58816-115), to remove spores from the sample matrix. Cold buffers (4°C) were used to minimize the potential for spore germination, prior to incubation, that could contribute to a PCR response at T0. Fourteen milliliters were then transferred to a 50 mL tube, and a second spore recovery step was conducted by addition of 14 mL cold Extraction Buffer without Tween (70% of 0.25 mM KH2PO4 (pH 7.4) and 30% ethanol). After brief vortexing, the second extraction volume was combined with the first to give 28 mL total volume. This allowed sufficient volume for equally splitting the sample between RV-PCR and culture analysis methods. Although 28 mL total were collected, only 13 mL were processed for culture or RV-PCR to avoid particles at the tube bottom that could interfere with accurate analysis.
After mixing, 13 mL of the suspension were transferred to a filter cup (Whatman Autocup™, GE Healthcare, Piscataway, NJ), and spores were collected on a 0.45 µm filter using a vacuum manifold and a vacuum pump (at 5-10 psi). The remaining spore extract was processed by traditional culture analysis described in the following section. After filtration, filter cups were washed with 20 mL of cold, 210 mM KH2PO4 buffer (pH 6.0), followed by washing with 7 mL of cold, 25 mM KH2PO4 buffer (pH 7.4). Filter cups were sealed on the bottom, 3.5 mL cold TSB medium were added, and top caps were added. The filter cup manifold was vortex mixed for 10 min, after which 1 mL T0 aliquots were withdrawn from filter cups for processing by PMPbased DNA extraction and purification [12], followed by real-time PCR analysis (see section below). The filter cups were sealed on the top and incubated for 9 hr (T9) at 35°C and 230 rpm. At T9, another 1 mL aliquot was withdrawn and processed, as described for the T0 aliquot. In addition, 100 µL aliquots at T0 and T9 were processed using a heat lysis protocol (including incubation at 95°C for 10 min). Heat lysis DNA extraction was performed in parallel for all samples as another check on performance of the PMP-based DNA extraction protocol. Results from heat lysis DNA extraction are only reported where there was a discrepancy between results from RV-PCR analysis using PMP-based DNA extraction and culture analysis; however, it should be noted that in most cases, the PMP-based method gave lower T9 Ct values than the heat lysis method, especially for samples with a high debris loading.
Culture sample processing and analysis
The remaining suspension was used for culture analysis as shown in Figure 1. Briefly, 13 mL were transferred to a second 50 mL conical tube, followed by centrifugation (3,184×g or 4,000 rpm for 30 min at 4°C) on an Eppendorf 5810R centrifuge (Eppendorf AG, Hamburg, Germany), to pellet the spores. After centrifugation, 9.5 mL of the supernatant were removed to give the same concentration factor, as that obtained for RV-PCR analysis (i.e. 13 mL filtered and resuspended in 3.5 mL on the filter cup). The pellet was then suspended and used to generate a serial dilution for plating. Ten-fold and 100-fold dilutions, as well as the undiluted spore suspension, were cultured on TSA plates in triplicate, and incubated 18-24 hr at 35°C. In addition, 0.5 mL of undiluted spore suspension was collected onto a filter membrane using a filter funnel apparatus (Pall Corp., Port Washington, NY, Cat. No. 4800), with the resulting filter placed onto a TSA plate for incubation (18-24 hr, 35°C). Finally, the remaining undiluted spore suspension was centrifuged, the supernatant was removed, and the pellet was suspended in 5 mL TSB to create an enrichment culture; the culture was incubated for 18-24 hr, at 35°C, and 200 rpm.
BG colony counts were obtained the next day based on colony morphology and the presence of orange pigmentation. Colony counts were corrected for dilution, in order to determine CFU/mL and CFU/ sample. If CFU were not evident and enrichment cultures were turbid, 10 µL aliquots were sampled by sterile inoculation loop and streaked for isolated BG colonies onto TSA plates (18-24 hr, 35°C). For each sample with presumptive BG colonies and/or a turbid enrichment culture, BG real-time PCR analysis of colonies (two from each sample if available), or the enrichment culture was used (after 10-fold concentration by centrifugation), to confirm the culture results. A DNA extraction protocol, which included heating at 95°C for 10 min, followed by centrifugation and removal of the supernatant for subsequent analysis was used. Five microliter aliquots were analyzed by the BG real-time PCR assay, and Ct values less than 35 were used to confirm that the sample had contained viable BG spores.
Additional culture analysis: concentration of the enrichment culture
Additional culture analysis was conducted in cases where positive RV-PCR results (ΔCt = 6 and Ct(T9) = 39), and/or Ct(T9) values less than 45 were obtained, but initial culture results were negative (no BG colonies were evident, and/or results were negative from initial real-time PCR analysis of the enrichment culture). Additional sample processing and analysis was performed in order to provide more accurate data for comparison of the two methods, including a more accurate assessment of the false positive and false negative percentages. The protocol included steps to analyze the enrichment culture using a rapid boil DNA extraction method, in the event that colonies were not evident (and therefore, could not be confirmed by real-time PCR). Since the original protocol did not include concentration of the enrichment culture prior to analysis, this represented only about 1/50th of the total enrichment culture volume processed (100 µL of the total 5 mL culture), and only about 1/20th of the crude DNA extract volume was analyzed by PCR (5 µL of the total 100 µL). In this study, 1 mL of culture was concentrated by centrifugation, with resuspension of the pellet in 0.1 mL (representing a concentration factor of 10), however, in some cases, negative BG real-time PCR results were still obtained. Therefore, for samples showing discrepancies between methods, the remaining enrichment culture was harvested by repeated centrifugation (1 mL aliquots), and the resulting pellet was suspended in 1 mL of Tris buffer (10 mM, pH 8) and processed using the 1 mL PMP-based DNA extraction protocol described above. The protocol provided an additional concentration factor of 1/5th, and also produced a cleaner DNA extract than that produced by the rapid boil protocol.
BG DNA standards for real-time PCR
The DNA standards were generated for the CRP BG strain. DNA was extracted from cultured cells (from spore stock outgrowth), using a MasterPure™ Complete DNA and RNA Purification Kit (Epicentre® Biotechnologies, Inc., Madison, WI), followed by RNase treatment. The DNA concentration was measured with a Qubit™ fluorometer, using the PicoGreen™ assay (Invitrogen™, Quant-iT™ dsDNA HS assay kit for Qubit fluorometer, Carlsbad, CA, Cat. No. Q32854). Standard concentrations were prepared in PCR-grade water. On each sample PCR plate, three replicates of 1 ng of BG genomic DNA per 25 µL PCR, three replicates of 100 fg of BG genomic DNA per 25 µL PCR, and one no-template control were included.
BG real-time PCR analysis
Five microliter sample aliquots were transferred to a 96-well Optical Fast plate with 20 µL of PCR mix. The BG real-time PCR assay targeted the recF gene, produced a 63-bp amplicon, and included the following components [3]: forward primer, Bg42F, with sequence 5’-CGCGCCCGAGGACTTAA-3’; reverse primer, Bg104R with sequence 5’-ATGTCAAGAAACCGCCGTC-3’; and fluorogenic probe, Bg60F/BHQ1: FAM-TCTCGTAAAGGGCAGCCCGCAAGBHQ1 (FAM, 6-carboxyfluorescein; BHQ, Black Hole Quencher®). PCR mix was prepared for the BG primer-probe set as follows: 12.5 µL TaqMan™ 2X Universal Master Mix (Applied Biosystems, Inc., Cat. No. 4305719), 1.25 µL each of forward and reverse primers, 2.5 µL probe, and 2.5 µL PCR water. After mixing and centrifugation, PCR was run using the ABI 7500 Fast platform (Applied Biosystems). The PCR thermal cycling parameters were as follows: 2 min at 50°C for uracil-N-glycosylase (UNG) incubation, 10 min at 95°C for AmpliTaq Gold® activation, followed by 45 amplification cycles (5 s at 95°C for denaturation and 20 s at 60°C for annealing/extension). For RV-PCR, each sample was analyzed in triplicate using the BG primer/probe set. The ROX reference dye contained in the 2X Universal Master Mix was used to normalize the fluorescent reporter signal. Automated analysis settings (baseline and threshold) were used throughout. If triplicate PCR results were not consistent (e.g. 1 of 3 or 2 of 3 positive), PCR was repeated until consistent results were obtained. T0 and T9 aliquots from the same sample were run on the same plate, in order to more accurately determine the ΔCt values.
Data interpretation and reporting
Initial and final PCR cycle thresholds Ct(T0) and Ct(T9), respectively, were used in the algorithm to determine whether viable (live) spores were present in the sample. Average values from triplicate analyses were used. Ct(T0) results that were non-detect (“Undetermined” with the PCR system software), were set to 45 in order to calculate a ΔCt value. For the BOTE project, a Ct(T9) of = 39 with a ΔCt (Ct[T0]-Ct[T9]) = 6 were set as cut-off values for a positive detection of viable BG spores. The ΔCt = 6 criterion represented an increase in DNA concentration at T9 relative to detectable DNA at T0, if any, as a result of the presence of viable spores in the sample that germinated and propagated during the 9 hours of incubation in growth medium.
The presumptive BG CFU were determined based on colony morphology and orange pigmentation, and counts between 25 and 250 were recorded for serial dilution plates, and counts between 1 and 100 were recorded for filter membrane plates. For plates or filters containing more than 250 or 100 colonies, respectively, the number was recorded as too numerous to count (TNTC). Presence of BG colonies on any of the sample culture plates, including those that were TNTC represented a positive result (BG presence) for that sample. CFU values were corrected for the dilution factor and expressed as CFU/sample. Realtime PCR data obtained from selected BG colonies (two per sample), and/or the enrichment culture was also reported. Analysis of enrichment cultures was only conducted if no BG colonies were detected for serial dilution or filter membrane plates, and cultures were turbid. The false positive percentage was determined by dividing the number of samples with positive RV-PCR results, but negative culture results by the total number of samples for that treatment, and multiplying by one hundred. The false negative percentage was calculated by dividing the number of samples with negative RV-PCR results, but positive culture results by the total number of samples for that treatment, and multiplying by one hundred.
Excel® spreadsheets were generated to streamline sample analysis and reporting, including results from positive and negative PCR controls. Sample results by sample barcode were reported via Excel spreadsheet for each sample batch. Data reports consisted of both RV-PCR Ct values (T0 and T9) and plate counts corrected for sample dilution, as well as qualitative data (positive/negative), for both RVPCR and traditional culture analysis. Control results from field and laboratory blanks/controls were also included in the reports. Since the RV-PCR method is qualitative, the comparison between RV-PCR and the culture method was performed in terms of positive/negative (presence/absence) BG detection. However, average ΔCt values (with standard deviations) and average plate count data (CFU per sample) were also reported.
Results and Discussion
Preliminary testing of buffers and spore preparation
Preliminary testing of buffers with and without ethanol to determine if ethanol impacted culture results showed there was no statistical difference between treatments; average CFU/mL from six replicates (corrected for dilution) showed 31,200 ± 1,700 CFU/mL for buffer with ethanol, compared to 29,400 ± 2,300 CFU/mL for buffer without ethanol. Subsequent analysis of modified extraction buffers with 2-fold lower Tween-80 for the first extraction step and no Tween-80 for the second spore extraction step, compared to the standard extraction buffer containing 0.1% Tween-80 (used for both extraction steps) showed no statistical differences between treatments (p = 0.09; Student’s t-Test, 2-tailed, paired), with average CFU/wipe (corrected for dilution) values of 9.99 ± 0.86 × 104 and 9.34 ± 0.45 × 104, respectively; therefore, slight changes in buffer composition to accommodate both culture and RVPCR analysis from the same sample did not impact BG spore recovery.
Preliminary experiments to characterize the CRP BG spore preparation showed that the material contained approximately 3.4×1011 CFU/g. Experiments conducted to characterize the spore preparation by direct PCR analysis of the spore suspensions showed that the 10-1 (104 mg/mL) and 10-2 spore dilutions did not show any Ct values (data not shown), likely due to inhibition since particulate material could be observed in the 10-1 dilution. In addition, the results for the 10-3 and 10-4 dilutions suggested there was partial inhibition, since the PCR data were not linear with the PCR data for the 10-5 through 10-9 dilutions (Table 1). For the 10-9 dilution (approx. 360 CFU/mL), less than one spore was estimated to be present for the PCR (5 µL aliquot), suggesting that extraneous cell debris with DNA, in addition to DNA on the spore coat, could contribute to the PCR response (Ave. Ct=28.5).
| Spore stock dilution | Estimated CFU in PCR | Average Ct (SD)* |
|---|---|---|
| 10-3 dilution (108/mL level) | 5 x 105 | 17.5 (0.4) |
| 10-4 dilution (107/mL level) | 5 x 104 | 21.5 (0.2) |
| 10-5 dilution (106/mL level) | 5 x 103 | 18.3 (0.2) |
| 10-6 dilution (105/mL level) | 5 x 102 | 21.5 (0.5) |
| 10-7 dilution (104/mL level) | 5 x 101 | 25.4 (0.3) |
| 10-8 dilution (103/mL level) | 5 | 28.8 (0.5) |
| 10-9 dilution (102/mL level) | 0.5 | 34.1 (2.3) |
*Averages and standard deviations are based on 3 replicates.
Acronyms: CFU, colony-forming units; Ct, cycle threshold; SD, standard deviation.
Table 1: BG real-time PCR assay results for CRP BG spore dilutions added directly to the reaction.
Although these results suggested a potential DNA background issue for use of the CRP BG spores with the RV-PCR protocol, when these spore dilutions were processed and analyzed by RV-PCR, no Ct (T0) responses were noted for any of the dilutions tested, including the 10-3 dilution (108 CFU/mL level) (Table 2). Furthermore, the Ct (T9) values were in the range expected from similar levels of vendorgenerated B. atrophaeus spore suspensions from previous experiments. In addition, an attempt was made to test the RV-PCR protocol on dead CRP BG spores. Spores for the 10-6 dilution were autoclaved 90 min at 126°C at 15 psi before spiking onto wipe samples; however, plating from the autoclaved spore suspension showed that an average of 12 CFU/mL remained after autoclaving. Results for this treatment were consistent with the 10-10 dilution, which had a similar log CFU level (Table 2).
| Spore Dilution | CFU/mL1 | CFU/filter cup2 | T0 Ct3 | Ave (SD) T9 Ct | Ave. ΔCt |
|---|---|---|---|---|---|
| 10-3 | 3.6E8 | 1.4E7 | NDT | 24.1 (1.0) | 20.9 |
| 10-4 | 3.6E7 | 1.4E6 | NDT | 24.8 (1.5) | 20.2 |
| 10-5 | 3.6E6 | 1.4E5 | NDT | 25.7 (0.6) | 19.3 |
| 10-6 | 3.6E5 | 1.4E4 | NDT | 27.0 (2.2) | 18.0 |
| 10-9 | 360 | 14 | NDT | 28.5 (1.4) | 16.5 |
| 10-10 | 36 | 1.4 | NDT | 33.1 (1.7) | 11.9 |
| 10-6 Killed4 | 12 | 4.7 | NDT | 34.4 (0.6) | 10.6 |
*The RV-PCR protocol was according to that described in the Materials and
Methods section except that 13 mL instead of 14 mL extraction buffer were
removed at each extraction step.
1The CFU/mL value was determined from plate counts of the CRP BG spore dilutions.
In each case, 100 μL of the appropriate spore solution was added to the wipe.
2The CFU/filter cup values were calculated by dividing the CFU/mL by 10 since 100
μL was added, then dividing by 33 since the total extraction buffer volume was 33
mL, and finally, multiplying by 13, since 13 mL of extraction buffer was added to
the filter cup.
3NDT = Non-detect; The Ct (T0) was set to 45 to calculate ΔCt.
4Plate counts showed that complete kill was not achieved.
Table 2: Summary of T0 Ct, T9 Ct, and ΔCt results for CRP BG spore dilutions spiked onto dirty wipes (250 mg autoclaved AZ Test Dust) and processed using the manual RV-PCR protocol*
Overall, these results showed that the RV-PCR protocol performed well even for relatively crude spore preparations. It appeared that the CRP BG spore material contained high levels of BG DNA not tightly associated with the spore coat, but readily removed during the RVPCR sample processing steps (spore extraction from the wipe and washes of the spores collected on the filter cup); therefore, the analysis could accurately distinguish live spores from spores merely containing associated DNA. Furthermore, RV-PCR results for T9 showed expected results suggesting the endpoint was sufficient to maintain the 10-spore level LOD.
BOTE project sample analysis
A total of 159 post-decontamination samples (surface wipe samples and QC samples) were analyzed during the BOTE project for the three different disinfection methods (Table 3). The QC samples were opened during sampling, but not used to sample surfaces. The distribution of QC and surface samples was not known before the sample analysis, although the total number per event was verified. In addition to the samples from INL, 12 TBs and 9 each of NCs and PCs were also analyzed with the post-decontamination samples. The TBs were sterile, pre-wetted wipes prepared in the same manner and at the same time as the wipes sent to the field team to use for sampling. The TB tubes were not opened, until they were processed for RV-PCR analysis. One PC, one NC, and at least one TB were included, with up to 21 samples on each tube rack/manifold. Samples were processed on the receipt date unless there was a shipment change from the original schedule, and then samples were stored at 4°C prior to processing. Up to 48 samples and controls were processed on the same day through both RV-PCR and traditional culture analyses. For some events, sample analysis (i.e. Post-CD samples) was split into two days, in order to accommodate the multiple steps required for culture analysis; however, the same sample was processed concurrently by RV-PCR and culture to enable direct comparison of results.
| BOTE Project Event1 | Sample Type2 | Number of Samples | Samples/Event | Total Samples |
|---|---|---|---|---|
| VHP | QC | 3 | 46 | 159 |
| Surface Sample | 41 | |||
| Bleach | QC | 8 | 67 | |
| Surface Sample | 39 | |||
| CD | QC | 8 | 84 | |
| Surface Sample | 60 |
1VHP, vaporous hydrogen peroxide; Bleach, pH-adjusted bleach; CD, chlorine
dioxide.
2QC, quality control (see text for description).
Table 3: Summary of post-decontamination samples received and processed by event type.
Results from sample analysis showed that pre-decontamination samples for the different treatments typically had 104-105 CFU per sample via culture analysis (data not shown). Lower viable counts (<101- 102 CFU per sample) were occasionally observed, which represented QC (field blank) samples rather than actual surface samples, although the sample type was not made known until after the data analysis was completed.
Samples from the vaporous hydrogen peroxide decontamination event
For Post-VHP samples, 41 of 44 (93%) were consistent between culture and RV-PCR analyses (Table 4), including three samples that met the algorithm for positive detection based on heat lysis results, whereas the ΔCt values were < 6 for the PMP-based DNA extraction (as noted). Eleven samples showed CFU that ranged from 101-103 CFU per sample. Several samples did not show CFU on the TSA plates for culture analysis, but were positive by real-time PCR analysis of the concentrated enrichment culture. Many of these were positive by RVPCR, however, while two samples showed a PCR response, they did not meet the criteria for positive detection by RV-PCR, namely Ct(T9) = 39 and ΔCt [Ct(T0)-Ct(T9)] = 6 (e.g. samples had ΔCt values of 5.6 and 3.7). In addition, one of the samples was positive by RV-PCR, but could not be confirmed by culture analysis. Such discrepancies are expected since samples contained very low spore levels (no CFU measured on plates), and as previously mentioned, there are likely heterogeneities in partitioning spores, while splitting the extract for parallel culture and RV-PCR analyses (e.g. see Figure 3).
| Sample ID | Culture (24-48 hr) | RV-PCR (9 hr) | ||||||
|---|---|---|---|---|---|---|---|---|
| Average CFU/Sample | Source of Culture Result* | PCR of Culture (Ct)** | Culture Result (Pos/Neg) | Average ΔCt | Std Dev ΔCt | RV-PCR Results (Pos/Neg) | Notes | |
| 1185 | 0 | EC-PE | 25.2 | Pos | 7.1 | 1.8 | Pos | See Footnote (1) |
| 1198 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1199 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1243 | 3.0E1 | Plates | 15.7 | Pos | 6.5 | 0.9 | Pos | |
| 1246 | 3.0E1 | Plates | 13.7 | Pos | 6.5 | 0.9 | Pos | |
| 1255 | 0 | EC | 25.6 | Pos | 5.6 | 0.3 | Neg | RV-PCR Ave. ΔCt from 1:20 dilution |
| 1257 | 3.0E1 | Plates | 14.6 | Pos | 6.1 | 1.3 | Pos | |
| 1259 | 0 | EC | 21.9 | Pos | 0.0 | 0.0 | Neg | Heat Lysis Ave. ΔCt = 3.7 |
| 1261 | 0 | EC-PE | 25.8 | Pos | 7.8 | 1.2 | Pos | See Footnote (1) |
| 1290 | 0 | EC-PE | 23.8 | Pos | 8.6 | 0.6 | Pos | See Footnote (1) |
| 1291 | 0 | EC | 25.6 | Pos | 10.9 | 0.1 | Pos | See Footnote (2) |
| 1292 | 0 | EC | 25.5 | Pos | 12.9 | 0.2 | Pos | See Footnote (2) |
| 1298 | 1.3E3 | Plates, FF | 12.9 | Pos | 8.3 | 0.7 | Pos | |
| 1300 | 3.0E1 | Plates | 13.6 | Pos | 6.7 | 0.8 | Pos | |
| 1301 | 0 | EC | 25.4 | Pos | 4.2 | 2.0 | Pos | Ave. ΔCt from 1:20 dilution; Heat Lysis Ave. ΔCt = 8.4 |
| 1302 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1303 | 0 | EC | 35.0 | Pos | 9.6 | 2.2 | Pos | |
| 1304 | 0 | EC | 23.8 | Pos | 14.8 | 0.1 | Pos | See Footnote (2) |
| 1305 | 5.3E1 | Plates, FF | 13.3 | Pos | 20.2 | 1.8 | Pos | |
| 1306 | 1.8E1 | FF, RS | 14.5 | Pos | 16.0 | 1.7 | Pos | |
| 1475 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1476 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 2.9E5 | Plates | 14.0 | Pos | 23.6 | 0.1 | Pos | |
| 1307 | 0 | EC | 35.0 | Pos | 6.7 | 0.4 | Pos | |
| 1308 | 8.9E1 | Plates, FF | 13.6 | Pos | 7.9 | 0.6 | Pos | |
| 1309 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1310 | 0 | EC | 16.2 | Pos | 8.7 | 0.3 | Pos | |
| 1311 | 0 | N/A | Undetermined | Neg | 6.4 | 0.3 | Pos | RV-PCR Ave. ΔCt from 2 replicates (1:20 dilution) |
| 1312 | 0 | EC | 26.0 | Pos | 7.7 | 0.3 | Pos | |
| 1374 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1375 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1377 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1379 | 0 | EC-PE | 24.8 | Pos | 12.3 | 0.5 | Pos | See Footnote (3) |
| 1382 | 0 | EC-PE | 27.0 | Pos | 12.6 | 0.1 | Pos | See Footnote (3) |
| 1383 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 4.7E5 | Plates | 14.0 | Pos | 24.9 | 0.1 | Pos | |
| 1163 | 0 | EC | Undetermined | Neg | 0.0 | 0.0 | Neg | |
| 1164 | 0 | RS | 15.0 | Pos | 5.9 | 0.6 | Pos | See Footnote (4): Heat Lysis Ave. ΔCt = 8.3 |
| 1165 | 5.9E1 | Plates | 14.7 | Pos | 8.8 | 0.2 | Pos | |
| 1168 | 0 | RS | 14.9 | Pos | 6.1 | 0.4 | Pos | |
| 1171 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1177 | 0 | RS | 15.2 | Pos | 8.6 | 0.7 | Pos | |
| 1179 | 0 | RS | 14.7 | Pos | 4.9 | 1.0 | Pos | See Footnote (4): Heat Lysis Ave. ΔCt 8.7 |
| 1180 | 0 | FF, RS | 34.6 | Pos | 9.3 | 0.3 | Pos | |
| 1181 | 0 | EC | Undetermined | Neg | 0.0 | 0.0 | Neg | |
| 1182 | 3.0E1 | Plates, FF | 14.4 | Pos | 7.0 | 0.8 | Pos | |
| 1186 | 0 | EC | 19.9 | Pos | 7.5 | 0.8 | Pos | |
| 1210 | 3.6E1 | FF, EC | 14.1 | Pos | 6.9 | 2.0 | Pos | |
| 1464 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1465 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1466 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 3.4E5 | Plates | 13.5 | Pos | 20.6 | 0.1 | Pos | |
*Positive (pos) or negative (neg) culture determination based upon the serial dilution (plates), filter funnel (FF) plate, and/or enrichment culture (EC), re-streak (RS) from
enrichment culture or Promega-extracted concentrated EC (EC-PE). Positive culture results obtained if > 0 Bg colonies were present on serial dilution or filter membrane
plates, and/or Bg-positive PCR results were obtained (Ct < 35) from colonies and/or the EC or RS.
N/A = not applicable; No CFU for analysis and EC not turbid and/or no
growth from RS.
**Cycle threshold (Ct) values obtained from real-time PCR analysis of selected colonies and/or enrichment culture (EC). Positive RV-PCR result based upon average ΔCt≥
6 and T9 Ct ≤ 39.
Footnote (1): EC concentrated 10- to 20-fold and DNA prepared using Promega extraction prior to PCR analysis to confirm culture result (EC-PE).
Footnote (2): PCR repeated to confirm RV-PCR result; 1:20 dilution result shown.
Footnote (3): RV-PCR results based on repeated PCR analysis with 1:20 dilution (1:10 dilution showed PCR inhibition); EC concentrated 10- to 20-fold and DNA prepared
using Promega extraction prior to PCR to confirm culture result (EC-PE).
Footnote (4): Low spore level, post-decontamination sample; sample positive by heat lysis RV-PCR.
Abbreviations: PC, positive control; NC, negative control; TB, true blank.
Table 4: RV-PCR and culture results for VHP post-decontamination samples.
Samples from the pH-adjusted bleach decontamination event
For Post-Bleach samples, there was 100% consistency between culture and RV-PCR results, with 47 of 47 samples in agreement. Results are shown in Table 5. For these samples, three samples were positive and 44 samples were negative for both methods. Two of the positive samples were only detected by real-time PCR analysis of the enrichment culture, whereas RV-PCR results clearly showed positive results with ΔCt values of 6.9 and 7.5. Only one Post-Bleach sample showed 101 CFU on serial dilution plates, and the corresponding RVPCR ΔCt value was 9.0, showing positive detection. One negative control sample showed positive results for culture analysis, likely due to the extra handling steps in the culture processing protocol, leading to cross-contamination; this sample was negative by RV-PCR analysis.
| Sample ID | Culture (24-48 hr) | RV-PCR (9 hr) | ||||||
|---|---|---|---|---|---|---|---|---|
| Average CFU/Sample | Source of Culture Result* | PCR of Culture (Ct)** | Culture Result (Pos/Neg) | Average ΔCt | Std Dev ΔCt | RV-PCR Results (Pos/Neg) | Notes | |
| 1260 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1314 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1315 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1316 | 1.8E1 | FF | 16.8 | Pos | 9.0 | 0.3 | Pos | See Footnote (1) |
| 1317 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1318 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1319 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1320 | 0 | EC-PE | 27.4 | Pos | 6.9 | 0.2 | Pos | See Footnote (2) |
| 1322 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1324 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1325 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1328 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1329 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1332 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1337 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1479 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 1.8E1 | Plates, FF | 15.3 | Pos | 0.0 | 0.0 | Neg | Cross-contamination for culture portion only |
| PC | 7.1E1 | Plates, FF | 15.1 | Pos | 11.4 | 0.3 | Pos | |
| 1341 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1342 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1344 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1346 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1348 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1349 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1352 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1354 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1355 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1356 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1357 | 0 | EC-PE | 25.0 | Pos | 7.5 | 0.3 | Pos | See Footnote (1) |
| 1358 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1359 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1360 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1361 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1480 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 8.9E1 | Plates, FF | 15.1 | Pos | 15.9 | 0.5 | Pos | |
| 1238 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1313 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1321 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1323 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1326 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1327 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1331 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1333 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1334 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1335 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1338 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1339 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1340 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1343 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1350 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1473 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 8.9E1 | Plates, FF | 15.6 | Pos | 10.1 | 0.1 | Pos | |
| 1353 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1362 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
*Positive (pos) or negative (neg) culture determination based upon the serial dilution (plates), filter funnel (FF) plate, and/or enrichment culture (EC).
Positive culture results obtained if > 0 Bg colonies were present on serial dilution or filter membrane plates, and/or Bg-positive PCR results were obtained (Ct < 35) from
colonies and/or the EC or RS.
N/A = not applicable; No CFU for analysis and EC not turbid and/or no growth from RS.
**Cycle threshold (Ct) values obtained from real-time PCR analysis of selected colonies and/or enrichment culture (EC).
Positive RV-PCR result based upon average ΔCt≥ 6 and T9 Ct ≤ 39.
Footnote (1): Culture-PCR result based on repeated analysis and 1:10 dilution.
Footnote (2): EC concentrated 10- to 20-fold and DNA prepared using Promega extraction prior to PCR, to confirm culture results (EC-PE).
Abbreviations: PC, positive control; NC, negative control; TB, true blank.
Table 5: RV-PCR and culture results for pH-adjusted bleach post-decontamination samples.
Samples from the chlorine dioxide decontamination event
For Post-CD samples, there was 100% consistency between culture and RV-PCR results, with 68 of 68 samples in agreement. Results presented in Table 6 show that all 68 samples were negative for both methods. In some cases, negative control samples were positive by culture analysis, while not for RV-PCR analysis, likely due to crosscontamination. Protocol modifications, including extra glove changes, were incorporated preventing this error in batches of culture analysis that followed.
| Sample ID | Culture (24-48 hr) | RV-PCR (9 hr) | ||||||
|---|---|---|---|---|---|---|---|---|
| Average CFU/Sample | Source of Culture Result* | PCR of Culture (Ct)** | Culture Result (Pos/Neg) | Average ΔCt | Std Dev ΔCt | RV-PCR Results (Pos/Neg) | Notes | |
| 1242 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1244 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1245 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1248 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1250 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1251 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1252 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1253 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1256 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1263 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1267 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1268 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1269 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1270 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1272 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1274 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1469 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 1.8E1 | Plates, FF | Undetermined | Pos | 0.0 | 0.0 | Neg | Cross-contamination observed for culture only |
| PC | 3.6E1 | Plates, FF | 29.7 | Pos | 12.3 | 0.1 | Pos | |
| 1281 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1282 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1284 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1286 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1330 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1336 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1402 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1403 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1404 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1406 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1407 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1409 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1410 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1411 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1412 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1415 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1470 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 1.2E2 | Plates, FF | 29.5 | Pos | 12.1 | 0.1 | Pos | See Footnote (1) |
| 1249 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1418 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1422 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1423 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1426 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1427 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1428 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1429 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1434 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1435 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1436 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1437 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1440 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1442 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1449 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1471 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 3.6E1 | Plates, FF | 15.5 | Pos | 10.2 | 0.3 | Pos | See Footnote (1) |
| 1450 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1390 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1391 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1392 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1393 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1398 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1399 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1400 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1401 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1405 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1439 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1441 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1443 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1444 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1472 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 5.3E1 | Plates, FF | 17.8 | Pos | 7.8 | 0.6 | Pos | |
| 1430 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1431 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1433 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1445 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1446 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1447 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1448 | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| 1474 (TB) | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| NC | 0 | N/A | N/A | Neg | 0.0 | 0.0 | Neg | |
| PC | 5.3E1 | Plates, FF | 15.6 | Pos | 7.1 | 0.1 | Pos | |
*Positive (pos) or negative (neg) culture determination based upon the serial dilution (plates) or filter funnel (FF) plate.
Positive culture results obtained if > 0 Bg colonies were present on serial dilution or filter membrane plates, and/or Bg-positive PCR results were obtained (Ct < 35) from colonies and/
or the EC or RS. N/A = not applicable; No CFU for analysis and EC not turbid and/or no growth from RS.
**Cycle threshold (Ct) values obtained from real-time PCR analysis of selected colonies and/or enrichment culture (EC).
Positive RV-PCR result based upon average ΔCt≥ 6 and T9 Ct ≤ 39.
Footnote (1): The PC results are from PCs prepared in different batches (spiked at either the 101 or 102 spores per wipe level).
Abbreviations: PC, positive control; NC, negative control; TB, true blank.
Table 6: RV-PCR and culture results for chlorine dioxide post-decontamination samples.
Summary and Conclusions
Overall, the RV-PCR method provided rapid results that were 98% consistent (156/159 post-decontamination samples) with results from culture analysis. The results are summarized in Table 7, including calculation of false positive and false negative percentages for RV-PCR based on differences with culture results. The overall false positive percentage for samples was 0.6% and overall false negative percentage was 1.4%. As discussed above, each sample was split into two equal parts (and concentrated to the same extent), and most of the samples showing discrepancies in results between methods represented samples, with low spore levels that did not generate CFU on culture plates whereas these samples were clearly positive by RV-PCR. There were seven cases where the culture result was only positive after concentrating the enrichment culture, followed by real-time PCR analysis; in most cases (except for two), the RV-PCR result was also positive for these samples. These results indicated that the RV-PCR could be more sensitive than the traditional plate culture methods, mainly due to the fact that it allows the use of the whole sample for the analysis.
| BOTE Project Event1 | Total Sample No.2 | Samples | True Blanks | Negative Controls | Positive Controls | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RV-PCR and Culture Pos. | RV-PCR and Culture Neg. | RV-PCR Pos. and Culture Neg. | RV-PCR Neg. and Culture Pos.3 |
Total Agreement4 (%) | False Pos.4 (%) | False Neg.4 (%) | RV-PCR and Culture Neg. | RV-PCR and Culture Neg.5 | RV-PCR and Culture Pos. | ||
| VHP | 44 | 30 | 11 | 1 | 2 | 41/44 (93.2%) |
1/44 (2.3%) |
2/44 (4.5%) |
5/5 | 3/3 | 3/3 |
| Bleach | 47 | 3 | 44 | 0 | 0 | 47/47 (100%) |
0/47 (0%) |
0/47 (0%) |
3/3 | 2/3 | 3/3 |
| CD | 68 | 0 | 68 | 0 | 0 | 68/68 (100%) |
0/68 (0%) |
0/68 (0%) |
5/5 | 4/5 | 5/5 |
| Total | 159 | 33 | 123 | 1 | 2 | 156/159 (98%) |
1/159 (0.6%) |
2/159 (1.4%) |
13/13 | 9/11 | 11/11 |
1VHP, vaporous hydrogen peroxide; Bleach, pH-adjusted bleach; CD, chlorine dioxide.
2Total sample number included surface and QC samples and did not include True Blanks or laboratory negative and positive controls.
3For two Post-VHP samples, the culture portion required PCR analysis of the concentrated enrichment culture to obtain positive results.
4Note that each sample was divided into two equal parts for parallel RV-PCR and culture analyses; as a result, variability could have been observed for the samples with
low spore levels. Percentages were based on surface and QC samples and did not include True Blanks or laboratory negative and positive controls.
5For two of eleven negative controls, culture results were positive but RV-PCR results were negative, suggesting cross-contamination occurred for the culture portion.
Abbreviations: Pos, Positive; Neg, Negative.
Table 7: Summary of post-decontamination sample viability analysis results by event type including blanks and controls.
In Tables 5 and 6, data from Post-Bleach and Post-CD were generated with some minor changes to the protocols, including washing aliquots in buffer prior to conducting DNA extraction (either by PMP-based or heat lysis protocols), using cold buffers and cold medium to prevent spore germination in T0 aliquots, and allowing coarser particles to settle out prior to performing liquid transfers. While changes improved the data quality, in some cases, the additional handling steps added to the risk of cross-contamination between samples that was manifested in some of the negative control samples, showing positive results for culture analysis (Table 7). To address the risk of additional handling steps, a more frequent glove change procedure was instituted, as well as changes in the final protocol to reduce the number of steps involving mixing of sample contents and settling. The mitigation measures appeared to be successful, given that cross-contamination was not evident in the subsequent sample processing efforts.
The high percent agreement between the two methods is noteworthy, given that the RV-PCR method had not previously been tested with post-decontamination field samples containing relevant levels of debris. The samples contained a wide range of spore levels (<101 to >105 CFU/ sample), with real-world debris loadings that were accurately detected by the RV-PCR method. The method evaluation was challenged by several samples containing low (<101 CFU/sample) to non-detect (by culture) spore levels in the background of nearly 106 dead spores. Since the method’s intended use is for post-decontamination clearance analysis, the high accuracy observed with the gold-standard culture method under relevant decontaminant scenarios provided a solid foundation for continued optimization and application of the method for virulent B. anthracis spores.
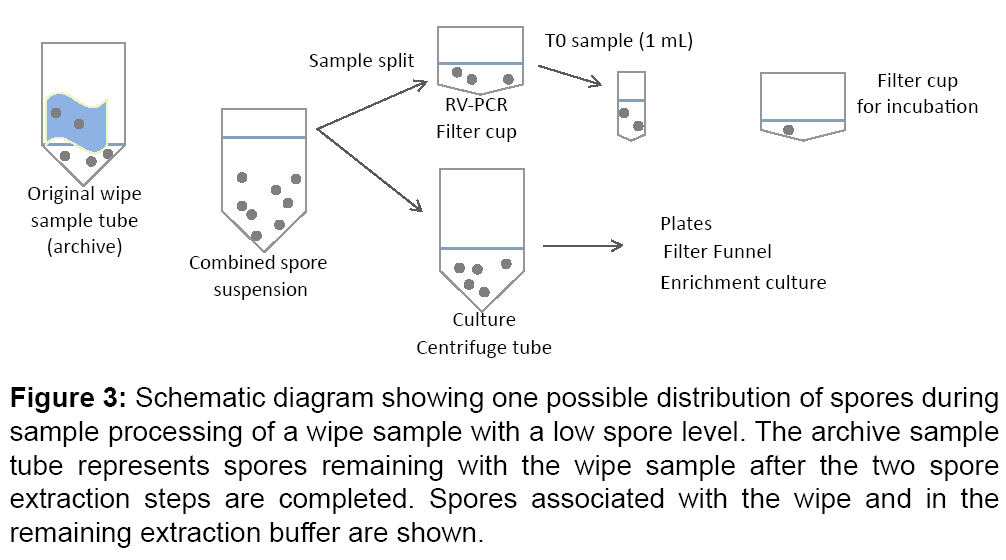
For low spore levels, more variability between culture and RVPCR results was expected due to factors such as spore clumping and pipetting variation for split sample extracts. A hypothetical spore distribution to different sample sub-sections is shown in Figure 3. In the combined protocol used for the BOTE project, the extract was split such that ~ 38% went to culture (13 mL), ~ 38% went to RV-PCR (13 mL) and ~ 24% remained unprocessed or archived (8 mL). At T0, 1 mL was removed for processing, which represented 28% of the total volume in the filter cup.
Figure 3: Schematic diagram showing one possible distribution of spores during sample processing of a wipe sample with a low spore level. The archive sample tube represents spores remaining with the wipe sample after the two spore extraction steps are completed. Spores associated with the wipe and in the remaining extraction buffer are shown.
As mentioned, samples also often contained high debris levels that provided a challenge for PCR-based analyses. However, RVPCR protocols were shown to be robust for environmental samples, and in most cases, the standard protocol gave consistent results with culture analysis. Occasionally, additional sample dilution (1 to 20 rather than 1 to 10 dilution) was required to obtain accurate RV-PCR results. In addition, the heat lysis DNA extraction protocol applied to a few samples showed greater ΔCt values than those based on PMPbased DNA extraction (Table 4). Further optimization of the DNA extraction and purification portion of the protocol is expected to address the observed PCR inhibition, as well as the higher apparent limit of detection for some post-decontamination samples. Protocol modifications introduced after the VHP sample testing (used for subsequent bleach and CD testing) also appeared to improve the PMPbased method, but this needs to be systematically evaluated, in order to ensure the accuracy of the RV-PCR approach across all sample types and relevant decontamination scenarios.
The method was shown to work well for BG spores exposed to decontaminants at real-world application levels, and with wipe samples containing background debris and indigenous microbial populations. The 98% agreement between methods was remarkable for a field test that included samples with low spore levels (below the detection limit of the plating method) after treatment with fumigants and surface disinfectants. The nine hour incubation endpoint appeared to be sufficient to detect any spores that might have been delayed in germination due to decontaminant exposure.
Based on 10-CFU level detection limits and solid performance with relevant challenges, the RV-PCR method has been included in EPA’s “Protocol for Detection of Bacillus anthracis in Environmental Samples During the Remediation Phase of an Anthrax Event” [13]. The method is also included with protocols for EPA’s Office of Emergency Management (OEM) Environmental Response Laboratory Network (ERLN), a network of laboratories for analysis of environmental samples following biological, chemical, or radiological attacks. The RV-PCR method may help meet the need for rapid, accurate viability analysis in the event of a B. anthracis release, thereby reducing the timeline for safely reopening facilities. Additional research is ongoing for RV-PCR applied to B. anthracis spores that were exposed to decontaminants, in order to confirm the robustness of the method for post-decontamination scenarios.
Acknowledgements
The support and guidance from the following EPA officials during the planning and execution of this effort are greatly appreciated: Dr. Hiba Ernst of the EPA National Homeland Security Research Center (NHSRC), and Dana Tulis and Erica Canzler of the EPA Office of Emergency Management (OEM). The support from the following persons for sample collection and shipment is sincerely acknowledged here: Dino Mattorano of the EPA-OEM, and Dr. Francisco Roberto and his team from the Idaho National Laboratory. We also sincerely thank all the members of the sampling teams from the EPA and the Civil Support Teams.
Dr. Gene Rice (EPA-NHSRC) and Dr. Stephanie Harris (EPA Region 10) are also acknowledged for their careful technical review of this manuscript, and for their suggestions. We sincerely thank Eletha Brady-Roberts for her thorough quality assurance review. We also thank Dr. Worth Calfee (EPA-NHSRC) for supplying stocks of CRP BG spores for preliminary testing, and for his guidance for wipe sample preparation.
Special thanks are extended to Romy Lee of the EPA-NHSRC for the help with the EPA-Department of Energy (DOE) Interagency Agreement management. The guidance from Thomas Bunt at LLNL is also greatly appreciated. Finally, we thank Dr. Joe Dalmasso from Yakibou Laboratories (formerly Apex Laboratories) for technical support with use of BG spore preparations.
This work was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory (LLNL), under Contract DE-AC52- 07NA27344. The United States Environmental Protection Agency through its Office of Research and Development and its Office of Emergency Management funded and managed the research described here (Interagency Agreement DW- 889-92328201-0). It has been subjected to LLNL and EPA administrative review and approved for publication. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States government or Lawrence Livermore National Security, LLC. The views and opinions expressed herein do not necessarily state or reflect those of the United States government or Lawrence Livermore National Security, LLC, and shall not be used for advertising or product endorsement purposes.
References
- Hodges LR, Rose LJ, O'Connell H, Arduino MJ (2010) National validation study of a swab protocol for the recovery of Bacillus anthracis spores from surfaces. J Microbiol Methods 81: 141-146.
- Rose LJ, Hodges L, O'Connell H, Noble-Wang J (2011) National validation study of a cellulose sponge wipe-processing method for use after sampling Bacillus anthracis spores from surfaces. Appl Environ Microbiol 77: 8355-8359.
- Kane SR, Létant SE, Murphy GA, Alfaro TM, Krauter PW, et al. (2009) Rapid, high-throughput, culture-based PCR methods to analyze samples for viable spores of Bacillus anthracis and its surrogates. J Microbiol Methods 76: 278-284.
- Létant SE, Kane SR, Murphy GA, Alfaro TM, Hodges LR, et al. (2010) Most-probable-number rapid viability PCR method to detect viable spores of Bacillus anthracis in swab samples. J Microbiol Methods 81: 200-202.
- Varughese EA, Wymer LJ, Haugland RA (2007) An integrated culture and real-time PCR method to assess viability of disinfectant treated Bacillus spores using robotics and the MPN quantification method. J Microbiol Methods 71: 66-70.
- Létant SE, Murphy GA, Alfaro TM, Avila JR, Kane SR, et al. (2011) Rapid-viability PCR method for detection of live, virulent Bacillus anthracis in environmental samples. Appl Environ Microbiol 77: 6570-6578.
- Nocker A, Camper AK (2006) Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol 72: 1997-2004.
- Rawsthorne H, Dock CN, Jaykus LA (2009) PCR-based method using propidium monoazide to distinguish viable from nonviable Bacillus subtilis spores. Appl Environ Microbiol 75: 2936-2939.
- Schmidlin M, Alt M, Brodmann P, Bagutti C (2010) Insufficient distinction between DNA from viable and nonviable Staphylococcus aureus cells in wipe-samples by use of propidium monazide-PCR. Appl Biosafety 15: 180-185.
- Shafaat HS, Ponce A (2006) Applications of a rapid endospore viability assay for monitoring UV inactivation and characterizing arctic ice cores. Appl Environ Microbiol 72: 6808-6814.
- Lee J, Deininger RA (2004) A rapid screening method for the detection of viable spores in powder using bioluminescence. Luminescence 19: 209-211.
- (2009) MagneSil(R) blood genomic, max yield system. Promega Technical Bulletin TB312, USA.
- USEPA (2012) Protocol for detection of Bacillus anthracis in Environmental samples during the remediation phase of an anthrax event. U.S. Environmental Protection Agency, Washington DC, USA.
Relevant Topics
- Anthrax Bioterrorism
- Bio surveilliance
- Biodefense
- Biohazards
- Biological Preparedness
- Biological Warfare
- Biological weapons
- Biorisk
- Bioterrorism
- Bioterrorism Agents
- Biothreat Agents
- Disease surveillance
- Emerging infectious disease
- Epidemiology of Breast Cancer
- Information Security
- Mass Prophylaxis
- Nuclear Terrorism
- Probabilistic risk assessment
- United States biological defense program
- Vaccines
Recommended Journals
Article Tools
Article Usage
- Total views: 14665
- [From(publication date):
specialissue-2013 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 9990
- PDF downloads : 4675