Research Article Open Access
Using Spatial Analysis of ANR1 Gene Transcription Rates for Detecting Nitrate Irregularities in Cherry Tomatoes (Solanum lycopersicum var. cerasiforme) Organic Greenhouses
Amir Mor-Mussery1,2*, Orit Edelbaum2, Arie Budovsky3, and Jiftah Ben Asher41The Department of Geography and Environmental Development, Ben Gurion University of the Negev, Bee’r Sheva, Israel
2TheDepartment of Soil and Water Sciences, Hebrew University, Rehovot, Israel
3Technological Center, Biotechnology Unit, Beer Sheva, Israel
4Katif R&D Center for Coastal Deserts Development Ministry of Science, Israel
- *Corresponding Author:
- Amir Mor-Mussery
Ben Gurion University of the Negev
Bee’r Sheva
Israel
Tel: 9720502213141
E-mail: Amir.mussery@gmail.com
Received date: January 15, 2017; Accepted date: June 27, 2017; Published date: July 04, 2017
Citation: Mor-Mussery A, Edelbaum O, Budovsky A, Ben Asher J (2017) Using Spatial Analysis of ANR1 Gene Transcription Rates for Detecting Nitrate Irregularities in Cherry Tomatoes (Solanum lycopersicum var. cerasiforme) Organic Greenhouses. Adv Crop Sci Tech 5: 286. doi: 10.4172/2329-8863.1000286
Copyright: © 2017 Mor-Mussery A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Organic fertilizers differ from the chemical ones by high inconsistency in their mineralization rates in the soil. This intrinsic property occasionally results in formation of distinct ‘soil patches’ different from one another with regard to the concentrations of soluble nutrients. This feature together with the different nutritional requirements during the crop’s physiological development complicates the designing of appropriate organic fertilization scheme. To overcome these difficulties a methodology was designed and tested based on analysis of molecular indicators. As a case study we tested cherry tomato (Solanum lycopersicum var. cerasiforme) organically grown in greenhouse located in Netzer Hazani village, Northern Negev, Israel at 2001. As a model nutrient we chose the soluble nitrate, due to its importance for the crop growth and development. We found that the best indicator of nitrate content were the ANR1 gene transcription rates which values were best correlated to the measured soluble nitrate in soil and the crop’s needs during development. Practically, the spatial analysis helped identifying the surpluses and deficits of soluble nitrate in soil patches which were subsequently treated in a quick and precise manner. Implementing this methodology on other crops and nutrients will allow constructing accurate and economical fertilization scheme which decrease the damages to ecosystems.
Keywords
ANR1 gene; Gene transcription rate; Soil soluble nutrients; Cherry tomatoes (Solanum lycopersicum var. cerasiforme); Spatial analyses in organic farming
Introduction
The organic farming requires use of fertilizers from organic sources such as compost, animal manure, animal urine, etc. [1]. Due to the complex structure of most of these fertilizers and the differences in their types, origin and composition, it is difficult to assess their mineralization rates. As a result, the organic fields are characterized by ‘areal soil patches’ with wide range in the levels of the soluble nutrients, as opposed to the homogeneity observed after chemical fertilization [2]. Therefore, there is a high chance that parts of the fields will have mineral content irregularities (deficits and surpluses) which could damage the crop growth and reproduction [3]. In the case of extreme surpluses, the minerals could even infiltrate to the ground water [4]. Due to this pattern of patches distribution, random sampling will not be useful for locating the irregular plots [5-7]. Additionally, the nutritional needs of the crop change during its physiological growth [8]. A summary for this state was given by Diacono and Montemurro [1] with relation to Nitrogen in their work entitled “Organic farming: challenge of timing Nitrogen availability to crop Nitrogen requirements” (for other nutrients see the following references: Potassium- Stockdale et al. [9]; Sulphur- Scherer [10]).
An alternative diagnosis to the soil nutrients analysis is based on the visual analysis of crop leaf state [11]. The biggest advantage of Wolf (1982) diagnosis is reflection of the plant nutrients needs in the tested physiological state. Still, this diagnosis suffers from inaccuracy and in many cases the results are lately achieved and the farmer cannot use them for planning his following practices. In the spirit of Wolf (1982) approach we measured the transcription rates of the nutrient dependent genes in the plant young leaves. As opposed to the plant’s DNA which remains un-changed during the plant life cycle (although claims have been made for possible changes in the chromosomes order, and even in the DNA sequences itself in higher plants in some cases; Walbot and Cullis [12], the mRNA amounts may change abruptly in response to the plant’s surrounding ecosystem (for changes in gene expression due to climate changes see- Lawlor and Mitchell [13], soil nutrients- Lambers et al. [14], and atmospheric gases composition and radiation- Willekens et al. [15], and even due to different soil practices- Park et al. [16]. In the recent years measuring of the mRNA levels has become a routine and rapid practice which takes only several minutes based upon use of commercially available low cost kits [17]. The analysis is based on sampling flag leaves which are characterized by rapid changes in molecular expression [18]. In the framework of the analysis, the cells walls of the flag leaves are broken [19], the mRNAs of the carrier proteins are processed [20], and the mRNA amounts of the examined genes is calculated (in our case, those related to the plant nutrition).
Thus, the aim of this study was to collect and analyze the transcription data from the field in order to locate fast the heterogeneous patches in organic sandy soil greenhouses based on the crop’s physiological stage.
Materials and Methods
Site of study
The study (an episode of long term study on growing organic crops in sandy soil and arid climate) was carried out in Netzer Hazani village, Northern Negev with rainfall of 400-500 mm year-1 (Longitude-15°34’, Latitude-20°31’). The upper soil horizon is based on dunes’ sand 10- 15 m thick [21], of uniform pattern (Particles content: Sand: 93%, Silt: 0.5% and Clay: 0%, Organic matter: 6.5%, pH: 7.9 and EC: 1.7, ‘Water holding capacity’: 30-40% and ‘Field capacity’: 10-35%). The site of study was greenhouse of 30 × 118 m size, planted with trellised Cherry tomatoes Solanum lycopersicum var. cerasiforme, type ‘495’ developed by Zeraim Gedera (distance between lines: 1 m and between the plants: 0.5 m, planting depth: 0.2 m). The seeds were sprouted and grown on organic bed for three weeks separately, and planted in the greenhouses at 15.02.2001. Prior to planting, 2.5 ton of ‘compost’ (the compost contains plant residues and animal manure, with relative ratio of 3:1, respectively), was mixed with the greenhouse soil at rate of 2.5 ton ha-1. The elements concentrations of the compost determined by Elemental analyzer EA1108 Eager [22] were as follows: N-5.53%, C-13.197%, H-6.24%, S-2.06%, C/N ratio of 1:3), Gaskell and Smith [23].
For maximizing the mRNA production in the crop leaves, despite the problematic aspects of the soil bed Tsoar [24], additional two organic fertilizers were added: During the growth season 0.15 Kg m-2 of Guano from Sunleaves© co. were added (Elements concentration from dry matter, based on supplier data is as follows: N-12%, P-11%, K-2%, www.sunleaves.com/Home/About).
Additionally, mount of 2 L was added to each plant (also called ‘cow urine’, composed of 95% water and 2.5% Urea, collected from cows breeder, Silva et al. [25] at planting and during the growth season, in order to assure that the essential nutritional elements were adequately supplied. The remaining agro technology implementation was based on organic farming protocols Baker [26].
As the key analyzed nutritional molecule, we chose the Soil Soluble Nitrate (SSNt) concentration due to the crops’ high Nitrate consumption rates from the soil by most of the crops as compared to the other nutritional compounds [27]. This choice was also evident in view of the tight correlation between the crops’ physiological development stage and their consumption rates of soil Nitrate [28]. Finally, the studied crop Cherry tomatoes (Solanum lycopersicum var. cerasiforme) was chosen due to its fast-growing rates, and rapid changes in different physiological stages [29]. For additional description of the agrotechnology see Mor- Mussery et al. [30].
Soil nitrate sampling and analysis
Soil samples, each weighing approximately 0.5 Kg, were taken in between the surface and 0.2 m depth at 07.04.2001. Overall, 27 samples were taken from the whole greenhouse in three sampling lines (distance of 10 m between the lines). The average distance between the samplings was 5 m. The extreme samples were taken 5 m from the edges to diminish the side effects. The soil samples were dried and mixed with distilled water (70 ml water per 100 gr of soil) for 24 hours. Afterwards, the soil solution was analyzed using spectrophotometer and compared to calibrated samples of known concentrations [22].
Plant tissue sampling and molecular analyses
From the whole greenhouse a representative area of 154 m2 (14 × 11 m with ~150 plants) was chosen (due to the research limitations) for the molecular analysis. From this area nine plants with the same physiological state were randomly marked based on gridding scheme, Miesch [31]. Immediately after the flower beds appearance (at 10.04.2001, eight weeks after planting, early in the morning when the genes’ transcription rates are minimally affected by sunlight radiation; Jansen et al. [32]) two flag leaves from each chosen plant were sampled, as suggested by Gregersen and Holm [33]. The leaves were frozen in liquid Nitrogen tank. The frozen tissues of the sampled plants were used for extracting the total RNA (RNATotal) using Quiagen® RNeasy mini kit (www.qiagen.com/ shop/sample-technologies/rna-sample-technologies/total rna/rneasymini- kit). Afterwards, analyses were performed in order to assess the RNATotal solutions purity. Primers known from scientific documentation to belong to eight genes associated with Nitrogen acquisition and assimilation were scanned on hybridization gel for identifying their binding specificity [34]. The primers themselves were synthesized by the authors using data from the Gene bank (www.ncbi.nlm.nih.gov). The genes were: ANR1 (gene related to roots elongation; Mathews et al. [35]); NRT1-2 (genes encoding for Nitrate transporter protein; Hildebrandt et al. [36]); Nia (gene which encodes the Nitrate reductase enzyme; Hildebrandt et al. [36]), and AMT1-3 (genes encoding ammonium transporter proteins; Von Wirén et al. [37]). The results of the gene specification analysis demonstrated that the synthesized primers of Nia (related to nitrate assimilation) and ANR1 (related to nitrate acquisition) genes demonstrated high specificity of hybridization, while the other primers bound to many unrelated genes. After this preliminary step, two analyses were performed - a PCR analysis for detecting the total amount of the Nia and ANR1 genes in each leave tissue, and Reverse Transcription PCRs (RT-PCR) for calculating the mRNA amounts related to these genes. The ratio between the mRNA and its DNA found in each examined tissue represents the Gene Transcription Rate (GTR), for further description see Spitzer-Rimon et al. [38].
Spatial analysis principles
The spatial analysis is based upon the assumption that plots in a given area have reciprocal influence on one another. As stressed earlier, in the field this influence manifests itself by the presence of areal soil patches. Specifically, for this study, the SSNt analysis, was performed as follows:
1. Measuring the soil’s Nitrate soluble concentrations and integrating it with the spatial locations.
2. Calculating the semi-variances of the Nitrate and graphing it against the separation distances of the measured plots - called ‘semi-variogram’.
3. The obtained semi-variances were used for calculating the Nitrate values all over the tested area based on ‘Simple Kriging’ interpolation [39]. Finally, the values were presented as distribution map divided into seven equal values groups, which allows locating the soil’s soluble nitrate irregularities (see also, Kravchenko et al. [40].
The described methodology was performed similarly for the Nia and ANR1 GTRs.
4. Crossing over the values maps of the SSNt and mRNAs concentrations belonging to different genes by using ‘Multilayer analysis’ allowed assessing the spatial interactions between them, and subsequently choosing the most reliable genes for the SSNt state [41].
*These analyses were performed by the ArcGIS Geostatistical Analyst®1 ver 10. and GS+® ver. 9.
*Any tool, equipment, fertilizer, software etc. mentioned in the paper do not reflect the authors’ preferences.
Results
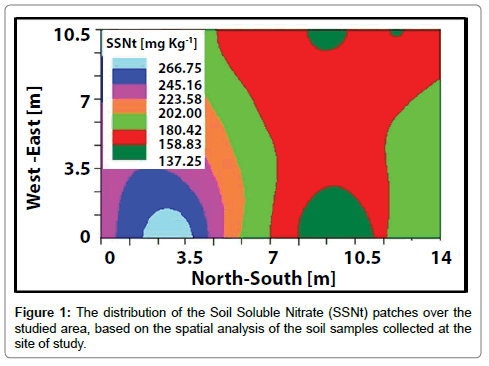
Figure 1 represents the SSNt concentrations distribution at the site of study after the spatial analysis. The patchiness pattern of the different SSNt concentrations is easily demonstrated with strips shape. Visual comparison of the calculated data from this study to the findings of Frias-Moreno et al. and Flores et al. [3,42] demonstrate that the SSNt concentrations of the 180-223 mg Kg-1 range are in the norm, which reflects more than 50% of variation from the studied area. The low values are mainly observed at the southern part, while the surpluses are found in the northern part.
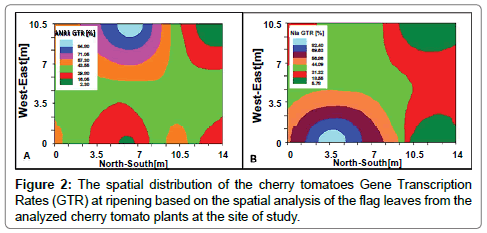
The Nia and ANR1 GTRs are demonstrated in Figure 2A and 2B, respectively. The GTRs highest values for ANR1 are in the west side of the tested area, while the highest ones for the Nia were in the eastern part. Inorder to get more precise correlation, a ‘pixel to pixel’ analysis (termed as ‘Multi-layer analysis’, Barbosa et al. [41]) was performed, by defining 7 SSNt value groups for both genes analyses.
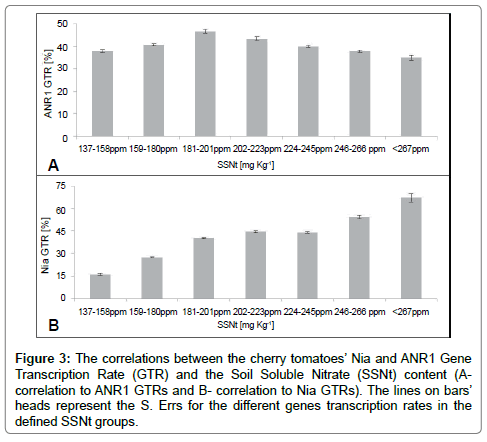
While the correlation between SSNt’ concentrations and the ANR1’s GTR could be defined by polynomial shape, with regard to the Nia we got linear shape (Figure 3B), both with regression coefficient of r2=0.97 (Figure 3A).
Figure 3: The correlations between the cherry tomatoes’ Nia and ANR1 Gene Transcription Rate (GTR) and the Soil Soluble Nitrate (SSNt) content (Acorrelation to ANR1 GTRs and B- correlation to Nia GTRs). The lines on bars’ heads represent the S. Errs for the different genes transcription rates in the defined SSNt groups.
Discussion
The significant correlations between the Nia and ANR1 transcription rates and SSNt concentrations demonstrate the usability of the described methodology in the case of Nitrate with relation to cherry tomatoes. This is true even in the case of organic management which is characterized by high heterogeneity in the soluble nutrient concentrations of the soil [2]. Crews and People [28] stressed that one of the biggest challenges of the present agriculture is “synchronizing the nitrogen supply and the crop demands”, so the next step should be identifying the most indicative gene (in this case either the Nia or the ANR1) of the plant’s SSNt needs. This data is valuable mostly before the ripening for planning immediate practices in order to get yield maximization (such as addition of cow urine or fish powder to correct SSNt deficits, CMG). In order to obtain description of the cherry tomatoes SSNt needs at ripening, we used Flores et al. data which define the norm for tomato crop between 180-220 ppm SSNt. In addition, Flores et al. [42] stressed that small deviations from this range could harm the plant’s development. Comparisons of the correlation patterns between the Nia and ANR1 GTRs, with the SSNt needs (Figure 3) as described by Flores et al. [42] revealed that the most indicative gene is the ANR1. Our observations could be explained by the tight correlation of the ANR1 expression levels to the fruit ripening Jaakola et al. [43] which makes the ANR1 gene more sensitive to irregularities. This is in contrast to the Nia GTR which has increased in the linear pattern even in the area of tomato surpluses which could be explained by its correlations to the vegetative growth of the tomatoes making it less sensitive to the SSNt surpluses (Frias-Moreno et al. [3].
For the farmer, the most important consideration of his work is the total costs and analysis duration. In Table 1 we compare the costs and durations of three methods for detecting the crop needs for nitrate in organic greenhouses. The first method is based on analyzing the total Nitrogen in the crop vegetative tissue. The second one is based on the soil soluble nitrate content, and the third one is based on “related nitrate” genes transcription rates (such as that of the ANR1 gene). It is easily demonstrable that mRNA analysis is not only the cheapest, but also the fastest method so the farmer can ‘repair’ different extremes in a fast manner.
| Total Nitrogen content analysis in vegetative tissues | Soil soluble nitrate content analysis | ANR1 transcription rate analysis with kit | |
|---|---|---|---|
| Field working | |||
| Duration | 1hour | 1hour | 1hour |
| Manpower* | 35$ | 35$ | 35$ (Iced CO2:15$) |
| Lab working | |||
| Duration | Tissues drying-2Dy, Extraction-1Dy,Analysis- 1Dy | Soil drying-1Dy, Extraction- 1Dy,Analysis-1Dy | 2hours2 |
| Costs | 800$(40$ per sample) | 600$ (40$ per sample)3 | Kit-110$(5.5$ per sample)1; Manpower- 40$ (20$ hour-1) |
| Spatial An. | Several minutes | Several minutes | Several minutes |
| Total (for 20 samples) | |||
| Total duration, costs | 4 Days,835$ | 3 Days,635$ | 3hours,200$ |
Table 1: Comparisons between different methods for spatially assessing the crop’s nitrate requirements based on common set for field study of 20 samples).
Still, several points have to be taken into account while discussing the findings of this study. The cherry tomato variety used by us (‘495’ from Zeraim Gadera’) is a newly introduced variety, and so the accurate data on the plant Nitrate needs were missing. In order to fill this gap, the authors used data from Frias-Moreno et al. [3] and Flores et al. [42] studies which were implemented on different varieties, so there is a reasonable chance that the actual SSNt needs of variety ‘495’ are different. Secondly, as Flores et al. stressed, there is an impact of the ratio between the Nitrate and Ammonia concentrations on the plant SSNt needs. Thirdly, the processing methodologies, soil characteristics, etc. have high effect on the SSNt needs (Frias-Moreno et al. [3]; Beaudoin et al. [44]).
Despite the above-mentioned reservations, future studies based on the principles described in this work will allow constructing comprehensive databases, containing for each crop the most indicative gene which mode of expression will be correlative with certain physiological stage of the crop such as vegetative growth, flourishing, ripening, and the reproductive stage [45]. The databases will also provide information on other factors having impact on the yield such as soil types, soil and irrigation, water salinity, and pH rates, so that the local farmers could adjust the relevant parameters for maximal production from their fields [46,47].
As stressed earlier, the costs of the mRNA extraction kits and subsequent analyses continue to decrease. Thus, the gene expression profiles could be obtained in a fast and easy manner [17].
Conclusions
We believe that by applying data regarding the crop soluble nutrients consumption amounts in a given physiological state, one can benefit from the gene transcriptional analysis presented here. This methodology is crucial mainly for nutrients characterized by difference between their fixed content and the soluble forms in the common fertilizers, as demonstrated here on Nitrate from organic source. The presented spatial analyses could be easily adjusted for many different fields such as greenhouses, or open areas under any processing scheme.
Acknowledgments
The paper is dedicated to Hilberg family from Nezer Hazani village for their extended help for implementing this research.
References
- Diacono M, Montemurro F (2010) Long-term effects of organic amendments on soil fertility. A review. Agron Sust Dev 30: 401-422.
- Tandon HLS(2005) Methods of analysis of soils, plants, waters, fertilizers and organic manures. Fertilizer Development and Consultation Organization.
- Frias-Moreno N, Nuñez-Barrios A, Perez-Leal R, Gonzalez-Franco AC, Hernandez-Rodriguez A,et al. (2014) Effect of Nitrogen deficiency and toxicity in two varieties of tomatoes (Lycopersicum esculentumL.). Agric Sci 5: 1361-1368.
- Addiscott TM, Whitmore AP, Powlson DS (1991) Farming, fertilizers and the nitrate problem. CAB International (CABI), p: 170.
- Heinze S, Raupp J, Joergensen RG(2010) Effects of fertilizer and spatial heterogeneity in soil pH on microbial biomass indices in a long-term field trial of organic agriculture. Plant Soil 328: 203-215.
- Watson CA, Atkinson D, Gosling P, Jackson LR, Rayns FW(2002) Managing soil fertility in organic farming systems. Soil Use and Management 18: 239-247.
- Pang XP, Letey J(2000) Organic farming challenge of timing nitrogen availability to crop nitrogen requirements. Soil Sci Soc America J 64: 247-253.
- Le Bot J, Adamowicz S, Robin P, Andriolo JL, Gary C(1997) Modelling nitrate uptake by greenhouse tomato crops at the short and long time scales. II. Modelling Plant Growth, Environmental Control and Farm Management in Protected Cultivation. ISHS Acta Hortic 456: 237-246.
- Stockdale EA, Shepherd MA, Fortune S, Cuttle SP(2002) Soil fertility in organic farming systems-fundamentally different?. Soil Use and Management 18:301-308.
- Scherer HW(2001) Sulphur in crop production-invited paper. Europ J Agron 14: 81-111.
- Wolf B(1982) A comprehensive system of leaf analyses and its use for diagnosing crop nutrient status. Comm Soil Sci Plant Anal 13:1035-1059.
- Walbot V, Cullis CA (1985) Rapid genomic change in higher plants. Ann Rev Plant Physiol 36: 367-396.
- Lawlor DW, Mitchell RA(2000) Crop ecosystem responses to climatic change: wheat. Climate change and global crop productivity, CAB International Press, pp:57-80.
- Lambers H, Raven JA, Shaver GR, Smith SE(2008) Plant nutrient-acquisition strategies change with soil age. Trends Ecol Evol 23: 95-103.
- Willekens H, Van Camp W, Van Montagu M, Inze D, Langebartels C, et al.(1994) Ozone, sulfur dioxide, and ultraviolet B have similar effects on mRNA accumulation of antioxidant genes in Nicotiana plumbaginifolia L. Plant Physiol 106: 1007-1014.
- Park W, Scheffler BE, Bauer PJ, Campbell BT (2012) Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.). BMC Plant Biology 12: 90.
- Rump LV, Asamoah B, Gonzalez-Escalona N(2010) Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from Salmonella cells. BMC Res Notes 3: 211.
- Zhou J, Wang X, Jiao Y, Qin Y, Liu X, et al. (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Molecular Biology 63: 591-608.
- Lin Y, Fuerst O, Granell M, Leblanc G, Lórenz-Fonfría V, et al. (2013) The substitution of Arg149 with Cys fixes the melibiose transporter in an inward-open conformation. Biochimica et Biophysica Acta (BBA)-Biomembranes 1828: 1690-1699.
- Lin Y, Bogdanov M, Tong S, Guan Z, Zheng L (2016) Substrate selectivity of lysophospholipid transporter LplT involved in membrane phospholipid remodeling in Escherichia coli. Journal of Biological Chemistry 291: 2136-2149.
- Ministry of the Environment Planning Department (1999) Coastal Zones Management in Israel.
- Eager (2001) Elemental Analyzer, Reference Manual, Eager Press, USA.
- Gaskell M, Smith R(2007) Nitrogen sources for organic vegetable crops. Hort Technol 17: 431-441.
- Tsoar H (1990) The ecological background, deterioration and reclamation of desert dune sand. Agric Ecosyst Environ 33:147-170.
- Silva RG, Cameron KC, Di HJ, Hendry T(1999) A lysimeter study of the impact of cow urine, dairy shed effluent, and nitrogen fertiliser on nitrate leaching. Aus J Soil Res 37: 357-370.
- Baker B (2009) Organic practice guide. Organic materials review institute.
- Ullrich WR(1992) Transport of nitrate and ammonium through plant membranes. In: Mengel K, Pilbeam DJ (eds.), Nitrogen Metabolism of Plants, pp: 121-137.
- Crews TE, Peoples MB (2005) Can the synchrony of nitrogen supply and crop demand be improved in legume and fertilizer-based agroecosystems? A review. Nutr Cycl Agroecosys 72: 101-120.
- Hobson G, Grierson D(1993) Tomato. Biochemistry of Fruit Ripening,pp: 405-442.
- Mor-Mussery A, Budovsky A, Ben-Asher J (2014) Geostatistical methods to evaluate the response of cherry tomato to soil nitrate. Int J Organic Agric Res Dev 9: 8-19.
- Miesch AT (1976) Methods of sampling, laboratory analysis, and statistical reduction of data. Geological Survey of Missouri.
- Jansen MA, Gaba V, Greenberg BM (1998) Higher plants and UV-B radiation: balancing damage, repair and acclimation. Trends in plant science 3: 131-135.
- Gregersen PL, Holm PB(2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotech J 5: 192-206.
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW(1995) Serial analysis of gene expression. Science 270: 484-487.
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, et al. (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. The Plant Cell 15: 1689-1703.
- Hildebrandt U, Schmelzer E, Bothe H(2002) Expression of nitrate transporter genes in tomato colonized by an arbuscular mycorrhizal fungus. Physiol Plant 115: 125-136 .
- Von Wirén N, Lauter FR, Ninnemann O, Gillissen B, Walch Liu P, et al. (2000) Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. The Plant J 21: 167-175.
- Spitzer-Rimon B, Marhevka E, Barkai O, Marton I, Edelbaum O, et al. (2010) EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles' biosynthesis in petunia. Plant Cell 22: 1961-1976.
- Bohling G (2005) Kriging Kensas Geology Press.
- Kravchenko AN, Bullock DG, Boast CW(2000) Joint multifractal analysis of crop yield and terrain slope. Agron J 92: 1279-1290.
- Barbosa JPRAD, Rambal S, Soares AM, Mouillot F, Nogueira JMP, et al.(2012) Plant physiological ecology and the global changes. Ciênciae Agrotecnologia 36: 253-269.
- Flores P, Carvajal M, Cerdá A, Martínez V(2001) Salinity and ammonium/nitrate interactions on tomato plant development, nutrition, and metabolites. J Plant Nutr 24: 1561-1573.
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, et al. (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153: 1619-1629.
- Beaudoin N, Saad JK, Van Laethem C, Machet JM, Maucorps J, et al.(2005) Nitrate leaching in intensive agriculture in Northern France: Effect of farming practices, soils and crop rotations. Agric Ecosys Environ 111: 292-310.
- Chaïb J, Devaux MF, Grotte MG, Robini K, Causse M, et al.(2007) Physiological relationships among physical, sensory, and morphological attributes of texture in tomato fruits. J Exp Bot 58: 1915-1925.
- Afzal M, Yousaf S, Reichenauer TG, Kuffner M, Sessitsch A (2011) Soil type affects plant colonization, activity and catabolic gene expression of inoculated bacterial strains during phytoremediation of diesel. J Hazard Mater 186: 1568-1575.
- Liu B, Mørkved PT, Frostegård Å, Bakken LR (2010) Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol Ecol 72: 407-417.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 2994
- [From(publication date):
June-2017 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 2187
- PDF downloads : 807