Research Article Open Access
Use of Bulk Segregant Analysis for Varietal Identification and Genetic Diversity Estimation in Pakistani Basmati and Non-Basmati Rice Cultivars through Molecular Markers
Arif M1*, Waheed R1, Muqaddasi QH1, Ahmed Z2, Khalid M4, Naveed SA1, Shahzad M1 and Xu J3
1DNA Markers and Applied Genomics Lab, Agriculture Biotechnology Division, National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan
2National Agriculture Research Centre, Islamabad, Pakistan
3Agricultural Genomics Institute at Shenzhen (AGIS), Chinese Academy of Agricultural Sciences (CAAS), China
4Government Emerson College, Multan, Pakistan
- Corresponding Author:
- Muhammad Arif
Genome Mapping Lab, Agriculture Biotechnology Division
National Institute for Biotechnology and Genetic Engineering (NIBGE)
Affiliated with PIEAS, Faisalabad, Pakistan
Tel: +92412651475/79
Fax: +92412651472
E-mail: marif_nibge@yahoo.com
Received date: April 07, 2016; Accepted date: May 25, 2016; Published date: June 05, 2016
Citation: Arif M, Waheed R, Muqaddasi QH, Ahmed Z, Khalid M, et al. (2016) Use of Bulk Segregant Analysis for Varietal Identification and Genetic Diversity Estimation in Pakistani Basmati and Non-Basmati Rice Cultivars through Molecular Markers. Adv Crop Sci Tech 4:227. doi:10.4172/2329-8863.1000227
Copyright: © 2016 Arif M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Identification of alleles for a particular variety through Bulk Segregant Analysis (BSA) is a valuable technique that probes differences specifically in distinguishing traits. Our study was an attempt to develop variety specific marker which differentiate aromatic rice cultivars from non-aromatic. Bulk Segregant Analysis (BSA) approach with RAPD and ISSR assay was undertaken to trace the variety specific alleles among 2 group of popular Pakistani Basmati and non-Basmati rice genotypes to check adulteration by rice exporters. Out of 160 RAPDs and 30 ISSR used in BSA, 29 RAPD and 18 ISSR revealed consistent polymorphisms and were used to assort the advanced disparities amongst 10 varieties. Overall 262 of 359 random amplified alleles and 116 of 151 inter simple sequence alleles were found polymorphic. The number of alleles generated from RAPD and ISSR marker ranged from 5 to 19 and 3-18 respectively. Pair-wise genetic similarity among the varieties with jaccard coefficients was 0.85 for RAPDs and 0.79 for ISSRs. UPGMA dendrogram based on cluster analysis of individual and combine genetic similarity co-efficient resolved 10 rice cultivars into 2 major groups, Basmati and Non-basmati. Moreover, a total of 3 variety specific alleles of 650, 800, 1075 bp were amplified by random primers OPN-05 and OPE-09. While ISSR generated five (UBC-814, UBC-811, UBC-808, UBC-808a and UBC-82) rare alleles for specific genotypes with band sizes of 1500, 1400, 250, 950 and 720 bp respectively. Direct sequencing of rare allelic ampliï¬ed bands in Sequence Characterized Ampliï¬ed Regions (SCARs) and sequence tagged sites (STS) might behave useful for recreating phylogenic trees with aim of preserving the integrity of Basmati rice.
Keywords
Bulk segregant analysis; Basmati rice (Oryza sativa L.); Genetic diversity; RAPD; ISSR; Varietal identification
Introduction
In recent years, emergence of Asian aromatic rice as a profitable business and globalization of world trade has necessitated formulation of legal tools for rice cultivar protection, certification and registration. The characterization of rice germplasm is obligatory not only for the identification of various cultivars but also to find out genetic relationships among them. Pakistan is globally recognized for cultivating and exporting long-grain aromatic basmati rice and its land is legendary for the production of extremely delicious rice with special characters i.e., aroma, high paste and extra-long grain [1].
In rice exporting countries, adulteration of rice varieties is awfully common. Several techniques (paddy rice visual examination, chemical method and DNA based analysis) have been employed over the last few decades, targeted specifically to the elimination of this problem. As an important export item, proper inspection and sophistication in preserving rice seed quality is imperative which is possible through development of variety specific markers. In addition to it, being evolved from extremely selected lines basmati breeding lines have narrow genetic base contributing only to the grain quality, aroma and high yield [2]. Characterization of the genetic resources is therefore, instantly needed because the strength of the genetic diversity is a requisite for world food security and endurance of human civilization.
DNA markers are valuable tools to resolve the genetic structure of a rice collection and to interpret the evolutionary associations among different groups [3]. Several techniques have been employed for diversity assessment but Bulk Segregant Analysis (BSA) along with RAPD and ISSR markers have proven to be useful in genetic fingerprinting, genotype identification, tagging the important genes for aroma, cooked kernel elongation, amylose content etc. and gene mapping [4]. BSA approach has been utilized to increase narrower genetic variation since this approach engrosses screening of differences between 2 pooled DNA samples [5]. The combined use of RAPD [6] and ISSR markers [7] allows for a high level of genomic coverage as RAPD markers are potentially associated with functionally important loci [8] and ISSR markers amplify hypervariable non-coding regions [9]. RAPD and ISSR are multi locus profiling techniques able to distinguish genotypes below the species level, such as cultivars and clones, and have been used in numerous diversity studies [10]. Moreover, the advantage of random pairing of primers lies in increasing the variability of polymorphic fragments and also in reducing the cost of designing primers. RAPD and ISSR primers have been successfully used for cultivar analysis in a number of plant species including rice [11]. Present research work was conducted to assess genetic diversity among different Basmati and Non-Basmati varieties via BSA and to identify markers linked to specific commercial varieties via BSA approach. The information obtained will be useful for adulteration testing as well as variety improvement breeding programs by minimizing the risk of genetic erosion.
Materials and Methods
Plant material and DNA extraction
10 Indica type Pakistani rice genotypes from two groups i) Basmati (Super Basmati, Basmati-198, Basmati-2000, Basmati-385, Basmati- Pak and Shaheen Basmati) and ii) Non-basmati (Pk-386, Supri, Supra and Super Fine) were selected and grown in field of Agriculture Biotechnology Division, National Institute of Biotechnology and Genetic Engineering (NIBGE) Faisalabad, Pakistan (Table 1). Total genomic DNA of each genotype was extracted from fresh leaves by CTAB method [12] whereas, quality and quantity of DNA was checked on 1.2% agarose gel and by spectrophotometer, respectively. The extracted DNA was diluted to a final concentration of 50 ng μL-1 with 1 X TE buffer and stored at -20°C.
| Group | Local Name | Remarks |
|---|---|---|
| Basmati | Super Basmati | Strong aroma, extra long grain, excellent cooking quality but very late maturing |
| Basmati-198 | Moderately aromatic, long grain but short stature, late maturing | |
| Basmati-2000 | Strongly aromatic, extra long grain, excellent cooking quality High yielding, local variety | |
| Basmati-385 | Aromatic, long grain, excellent cooking quality after Super Basmati, short stature, early maturing, High yielding | |
| Basmati-Pak (Kernal Basmati) | Moderately aromatic, extra long grain, excellent cooking quality, local variety, late maturing, tall, High yielding, | |
| Shaheen Basmati | Moderately aromatic, extra long grain, excellent cooking quality, local variety | |
| Non- Basmati | PK-386 | High yielding, Non-aromatic, long grain, early maturing variety, poor cooking quality |
| Supri | High yielding, Non-aromatic, long grain, early maturing variety | |
| Supra | High yielding, Non-aromatic, long grain, early maturing variety | |
| Super fine | High yielding, Non-aromatic, long grain, early maturing variety |
Table 1: List of Rice varieties used for RAPD and ISSR analysis.
Bulk Seggregant RAPD and ISSR analysis
Two bulked DNA samples are generated to discriminate Basmati from Non-Basmati varieties. The bulks are screened for differences using RAPD assay. A total of 160 RAPD primers from Operon Technologies Inc, Alabama, USA series C, D, E, R and Gene Link Series A, B, M, N were used. RAPD amplification reaction were carried out in a 20 μL reaction containing 40 ng genomic DNA, 3 μL of primer, 100 μM dATP, dCTP, dGTP and dTTP, 1 unit of Taq polymerase (fermentas), 1 × Taq polymerase buffer and 2.5 mM MgCl2. DNA amplification reaction was performed for RAPD-PCR as: Initial denaturation of 5 min at 95°C, 40 cycles of 94°C for 1 min (denaturation), 37°C for 1 min (annealing), 72°C for 2 min and final extension at 72°C for 10 minutes according to Williams et al. [6]. Same samples were analyzed by 30 ISSR markers of which 18 showed good amplification and were used for analysis. Amplification was performed in a 25 μL reaction mixture containing 40 ng of genomic DNA, 0.3 μM each primer, 1 × Taq DNA polymerase reaction buffer, 1.5 U of Taq DNA polymerase and 0.2 mM each dNTPs [13]. Reproducibility of amplified RAPD and ISSR markers were tested for their ability to prime PCR amplification of three selected cultivars with independent amplifications from three independent DNA isolations per cultivar. For the choice of RAPD/ISSR primers, unambiguous and qualitative (present or absent) fragments that gave repeatable patterns two or three times with the same cultivar were considered. Primers that amplified consistently reproducible polymorphisms were selected and used to analyze all the 10 cultivars. The amplified products of both RAPD and ISSR were resolved on 1.2% agarose gels with ethidium bromide staining, photographed by UV light and scored for the presence and absence of DNA fragments. The PCR product was scored by means of polymorphic banding pattern.
Data analysis
The amplified products were scored for the presence (1) or absence (0) of bands of various sizes across the genotypes to generate a binary matrix. To quantify the genetic shift, a similarity matrix was generated from the binary data of RAPD and ISSR individually and combinely using Jaccard coefficients in program of XLSt at 2014. Cluster analysis, done with the UPGMA constructed dendrogram based on similarity co-efficient was used to assess pattern of genetic diversity among rice genotypes. After determining that each string of 1 and 0 scores was a unique array (each individual had a unique string of traits), all possible pair wise genetic distance values were calculated by Jaccard indexes [14]. Polymorphic information content (PIC) values were calculated for each RAPD and ISSR primer according to the formula: PIC=1-Σ(Pij)2, where Pij is the frequency of the ith pattern revealed by the jth primer summed across all patterns revealed by the primers [15]. Principal Coordinate analysis (PCoA) was performed using GenAlEx version 6.501.
Results
Bulk segregant analysis and cultivar identification
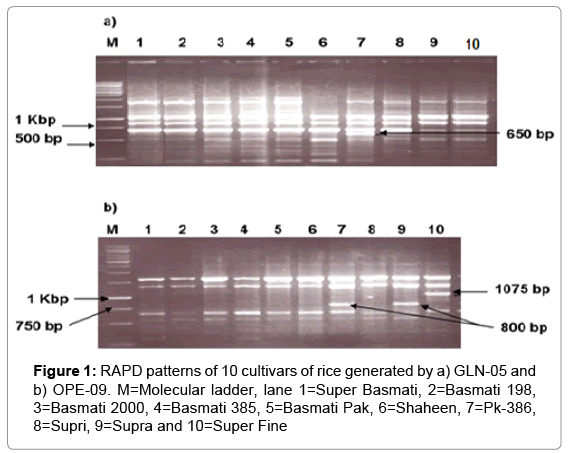
Among the tested markers, 55 primers were found to be polymorphic in bulk segregant RAPD analysis. Out of these markers, 29 showed lucid and consistent banding patterns for amplification of each cultivar were ultimately chosen for examining the genetic diversity and discrimination among the cultivars used (Table 2). While 18 polymorphic ISSR markers exhibited several bands that were shared among the basmati and other fine cultivars, whereas a few bands were obtained from some primers that revealed variety specific bands. Amplification profile revealed by two of the random polymorphic markers (OPN-05 and OPE-09) shows a characteristic variety specific band (Figure 1).
| Sr. No. | Primers | Sequence (5’-3’) | Number of loci | Polymorphism (%) | Maj. Allele freq. | Var. Spec. band | PIC | ||
|---|---|---|---|---|---|---|---|---|---|
| Total | Polymorphic | ||||||||
| RAPD primers | |||||||||
| 1 | GLN-05 | ACTGAACGCC | 14 | 12 | 85.7 | 0.2 | 1 | 0.8680 | |
| 2 | GLN-09 | TGCCGGCTTG | 9 | 6 | 66.6 | 0.4 | 2 | 0.7300 | |
| 3 | GLN-11 | TCGCCGCAAA | 14 | 10 | 71.4 | 0.2 | 0.8442 | ||
| 4 | GLN-12 | CACAGACACC | 13 | 10 | 76.9 | 0.3 | 0.7978 | ||
| 5 | GLN-14 | TCGTGCGGGT | 15 | 13 | 86.6 | 0.3 | 0.8232 | ||
| 6 | GLN-15 | CAGCGACTGT | 10 | 5 | 50 | 0.8 | 0.3142 | ||
| 7 | GLN-16 | AAGCGACCTG | 11 | 6 | 54.5 | 0.2 | 0.8196 | ||
| 8 | GLN-19 | GTCCGTACTG | 18 | 13 | 72.2 | 0.3 | 0.8232 | ||
| 9 | GLN-20 | GGTGCTCCGT | 11 | 7 | 63.6 | 0.3 | 0.7978 | ||
| 10 | GLA-04 | AATCGGGCTG | 10 | 5 | 50 | 0.4 | 0.7014 | ||
| 11 | GLA-17 | GACCGCTTGT | 14 | 11 | 78.5 | 0.1 | 0.8910 | ||
| 12 | GLB-01 | GTTTCGCTCC | 7 | 6 | 85.7 | 0.4 | 0.6756 | ||
| 13 | GLB-08 | GTCCACACGG | 12 | 10 | 83.3 | 0.2 | 0.8680 | ||
| 14 | OPC-04 | CCGCATCTAC | 8 | 2 | 25 | 0.4 | 0.5632 | ||
| 15 | OPD-04 | TCTGGTGAGG | 5 | 2 | 40 | 0.6 | 0.5700 | ||
| 16 | OPE-03 | CCAGATGCAC | 16 | 9 | 56.2 | 0.2 | 0.8442 | ||
| 17 | OPE-04 | GTGACATGCC | 13 | 11 | 84.6 | 0.3 | 0.7724 | ||
| 18 | OPE-06 | AAGACCCCTC | 7 | 6 | 85.7 | 0.2 | 0.8660 | ||
| 19 | OPE-09 | CTTCACCCGA | 7 | 3 | 42.8 | 0.6 | 0.5200 | ||
| 20 | GLM-02 | ACAACGCCTC | 13 | 11 | 84.6 | 0.3 | 0.8232 | ||
| 21 | GLM-03 | GGGGGATGAG | 14 | 10 | 71.4 | 0.3 | 0.7716 | ||
| 22 | GLM-04 | GGCGGTTGTC | 16 | 12 | 75 | 0.2 | 0.8680 | ||
| 23 | GLM-09 | GTCTTGCGGA | 15 | 14 | 93.3 | 0.1 | 0.8910 | ||
| 24 | GLM-11 | GTCCACTGTG | 15 | 13 | 86.6 | 0.2 | 0.8680 | ||
| 25 | GLM-13 | GGTGGTCAAG | 12 | 9 | 75 | 0.2 | 0.7754 | ||
| 26 | GLM-15 | GACCTACCAC | 14 | 7 | 50 | 0.3 | 0.7210 | ||
| 27 | GLM-16 | GTAACCAGCC | 9 | 6 | 66.6 | 0.3 | 0.7042 | ||
| 28 | GLM-18 | CACCATCCGT | 18 | 17 | 94.4 | 0.2 | 0.8823 | ||
| 29 | GLM-20 | AGGTCTTGGG | 19 | 16 | 84.2 | 0.2 | 0.6562 | ||
| Total | 359 | 262 | 8.7 | 3 | 22.05 | ||||
| Average | 12.4 | 9 | 70.4 | 0.3 | 1.5 | 0.760 | |||
| ISSR Primers | |||||||||
| 1 | UBC-807 | AGAGAGAGAGAGAGAGT | 11 | 6 | 54.5 | 0.4 | 0.7300 | ||
| 2 | UBC-808 | AGAGAGAGAGAGAGAGC | 18 | 11 | 61.1 | 0.1 | 1 | 0.8680 | |
| 3 | UBC-810 | AGAGAGAGAGAGAGAGT | 13 | 10 | 76.9 | 0.2 | 0.8910 | ||
| 4 | UBC-811 | GAGAGAGAGAGAGAGAC | 6 | 4 | 66.6 | 0.2 | 1 | 0.7942 | |
| 5 | UBC-814 | CTCTCTCTCTCTCTCTA | 6 | 5 | 83.3 | 0.5 | 2 | 0.6730 | |
| 6 | UBC-816 | CACACACACACACACAT | 6 | 5 | 83.3 | 0.6 | 0.6980 | ||
| 7 | UBC-820 | GTGTGTGTGTGTGTGTC | 11 | 10 | 90.9 | 0.1 | 1 | 0.8910 | |
| 8 | UBC-821 | GTGTGTGTGTGTGTGTC | 9 | 8 | 88.8 | 0.4 | 1 | 0.7578 | |
| 9 | UBC-824 | TCTCTCTCTCTCTCTCG | 7 | 6 | 85.7 | 0.2 | 0.8442 | ||
| 10 | UBC-826 | ACACACACACACACACC | 14 | 12 | 85.7 | 0.3 | 0.8232 | ||
| 11 | UBC-827 | ACACACACACACACACCG | 7 | 5 | 71.4 | 0.3 | 0.7978 | ||
| 12 | UBC-828 | TGTGTGTGTGTGTGTGA | 5 | 4 | 80.0 | 0.4 | 0.7300 | ||
| 13 | UBC-829 | TGTGTGTGTGTGTGTGC | 6 | 5 | 83.3 | 0.3 | 0.7716 | ||
| 14 | UBC-834a | AGAGAGAGAGAGAGAGCT | 5 | 3 | 60.0 | 0.8 | 0.2688 | ||
| 15 | UBC-834b | AGAGAGAGAGAGAGAGTT | 3 | 1 | 33.3 | 0.5 | 0.4918 | ||
| 16 | UBC-836a | AGAGAGAGAGAGAGAGCA | 7 | 4 | 57.1 | 0.3 | 0.7716 | ||
| 17 | UBC-836b | AGAGAGAGAGAGAGAGTA | 11 | 10 | 90.9 | 0.2 | 0.8442 | ||
| 18 | UBC-840 | GAGAGAGAGAGAGAGATT | 9 | 8 | 88.8 | 0.2 | 0.8680 | ||
| Total | 151 | 116 | 6 | 6 | 13.5 | ||||
| Average | 8.882 | 6.82 | 76.99 | 0.33 | 1.2 | 0.75 | |||
Table 2: RAPD and ISSR primers used to detect polymorphism, No. of polymorphic bands among different cultivars per primer.
Number of alleles and percentage polymorphism
The level of polymorphism among these rice cultivars was estimated by calculating allele number and polymorphism for each of the evaluated 29 RAPDs and 18 ISSR. A total of 262 polymorphic alleles were detected using these markers. The index of polymorphism, which was estimated as the proportion of variable random loci in the total number of loci studied, was 70.4% (average over all loci), varying from 25-94.4% depending on primers used (Table 2). The number of alleles per locus generated by each RAPD marker varied from 5 to 19 with an average of 12.4 alleles per locus. Maximum numbers of 17 polymorphic alleles were obtained with the marker GML-18, while the minimum 2 alleles/marker was amplified with OPC-04 and OPD-04, with an average of 9 polymorphic bands.
Eighteen ISSR primers showing reproducible and polymorphic patterns were chosen for cultivar identification and generated a total of 151 bands, with an average of 8.8 bands per primer. The size ranged from 250 to 3000 bp. Of the 151 bands produced, 116 (76.9%) were polymorphic. The number of polymorphic bands detected with each primer ranged from 3 (UBC834b) to 18 (primer UBC808) with an average of 6.82 per locus (Table 2).
Rare alleles
Alleles with a frequency less than 5%, were recognized as rare alleles. Generally, markers identifying a greater number of alleles per locus manifest more rare alleles. In our study, a total of 9 rare alleles were observed, 3 from 262 random polymorphic loci and 6 from ISSR loci. The cultivar Pk-386 with primer OPN-05 displayed a unique band of about 650 bp in comparison with all other cultivars. Interestingly, another primer OPE-09 revealed characteristic fragment of 1075 bp in Super Fine and 800 bp in each Supra and Pak-386 which were not produced in any of the other cultivars used (Figure 1). Among ISSR markers, five markers were cultivar specific and able to distinguish them clearly.
Similarity matrix and cluster analysis
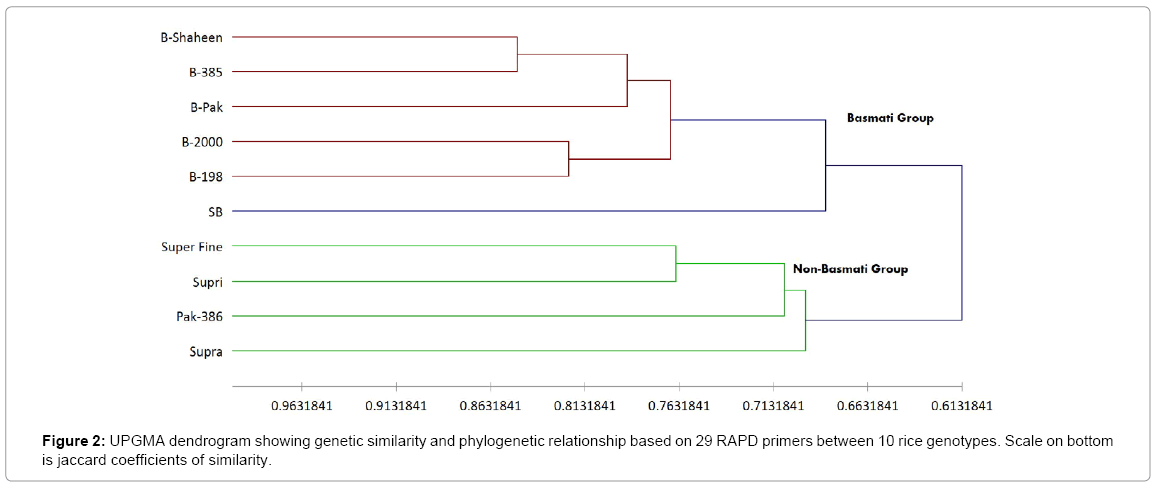
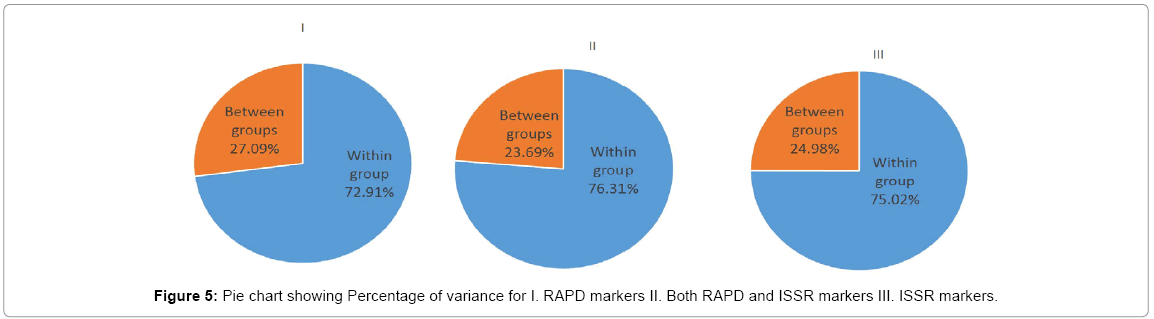
A similarity matrix based on the proportion of shared alleles between RAPD and ISSR was used to establish the level of relatedness between the cultivars surveyed (Table 3). Pair-wise genetic similarity coefficients by RAPD markers among these varieties varied from 0.527 to 0.849 with an average similarity of 0.67 (Table 3 lower diagonal). The genetic relationship among the rice cultivars was assessed by a cluster analysis of the similarity matrix. Dendrogram based on RAPD marker revealed that at about 0.61 similarity coefficients, the 10 rice genotypes grouped into two major clusters, which effectively differentiated the basmati from non-basmati cultivars (Figure 2). Landraces with aroma trait (Basmati-Shaheen, Basmati-385, Basmati-Pak, Basmati-2000, B-198 and Super Basmati) showed limited variability and clustered together in group-I at similarity co-efficients of 0.68 (68%). Group II included Non-Basmati, non-aromatic quality cultivars i.e., Supri, Super fine, Supra and Pak-386, which were clustered together at similarity coefficient of 69.6% (0.696). Maximum percentage of genetic variation was present within the groups i.e., 72.9% followed by 27% diffrences among cluster. This Variance among and within the populations are highly significant.
| SB | B- 198 | B-2000 | B- 385 | B-Pak | B-Shaheen | Pk-386 | Supri | Supra | Super Fine | |
|---|---|---|---|---|---|---|---|---|---|---|
| SB | **** | 0.638 | 0.752 | 0.722 | 0.821 | 0.613 | 0.584 | 0.403 | 0.561 | 0.588 |
| B-198 | 0.669 | **** | 0.604 | 0.604 | 0.600 | 0.532 | 0.514 | 0.417 | 0.546 | 0.482 |
| B-2000 | 0.688 | 0.822 | **** | 0.719 | 0.788 | 0.623 | 0.581 | 0.457 | 0.530 | 0.585 |
| B-385 | 0.678 | 0.738 | 0.810 | **** | 0.772 | 0.681 | 0.637 | 0.470 | 0.571 | 0.572 |
| B-Pak | 0.710 | 0.740 | 0.799 | 0.774 | **** | 0.633 | 0.661 | 0.435 | 0.569 | 0.634 |
| B-Shaheen | 0.683 | 0.743 | 0.777 | 0.849 | 0.807 | **** | 0.517 | 0.443 | 0.504 | 0.536 |
| B-386 | 0.573 | 0.623 | 0.642 | 0.593 | 0.653 | 0.668 | **** | 0.504 | 0.666 | 0.628 |
| Supri | 0.547 | 0.620 | 0.631 | 0.591 | 0.613 | 0.618 | 0.739 | **** | 0.490 | 0.583 |
| Supra | 0.572 | 0.673 | 0.659 | 0.640 | 0.654 | 0.621 | 0.678 | 0.716 | **** | 0.605 |
| Super Fine | 0.527 | 0.570 | 0.597 | 0.593 | 0.608 | 0.629 | 0.676 | 0.765 | 0.695 | **** |
Table 3: Genetic similarity coefficients of 10 rice genotypes based on RAPD (below diagonal) and ISSR (upper diagonal) markers.
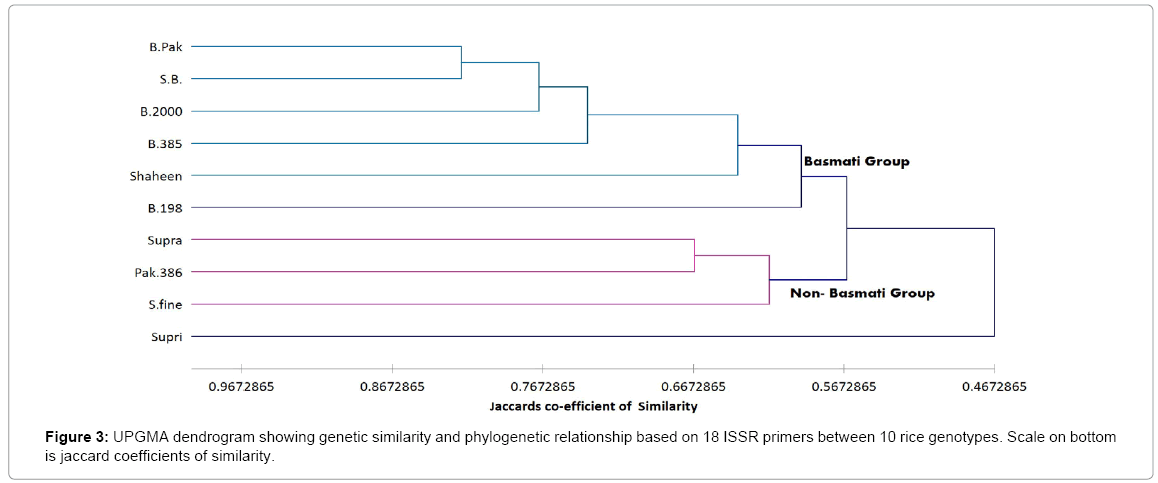
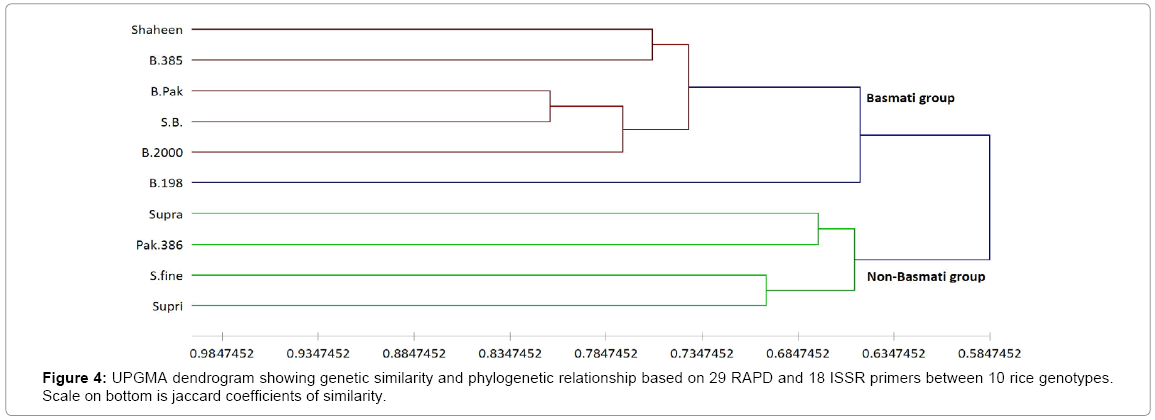
The ISSR genetic similarity values based on Jaccard’s coefficient ranged from 0.4 between ‘Supri’ and ‘SB’ to 0.89 between ‘B.000 and B. Pak (Table 3 upper diagonal). A dendrogram that was constructed according to ISSR data of ten rice cultivars allowed us to divide them into two main clusters (Figure 3). First cluster included all Basmati varieties i.e., Basmati-Shaheen, Basmati-385, Basmati-Pak, Basmati-2000, B-198 and second included all non-Basmati varieties i.e., Super fine, Supra and Pak-386 except Supri. The greatest similarity (78%). was observed between species B. Pak and B.2000 (Figure 3). A dendrogram constructed from combine data generated by RAPD and ISSR markers also clearly discriminate Basmati varieties from Non- Basmati group (Figure 4).
Principal Coordinate Analysis (PCoA)
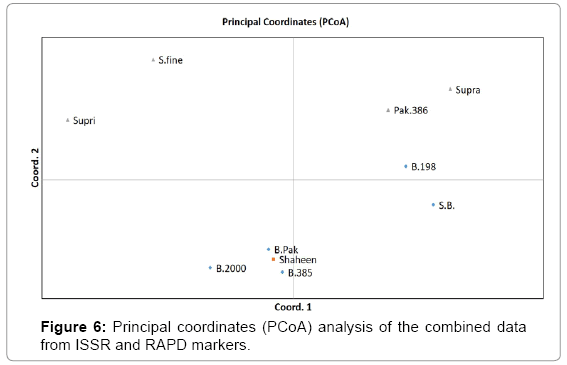
In Principal Coordinate Analysis (PCoA) the first three coordinates explained 64.2% of the total variation, 27.4% explained by the first coordinate with 22.1% and 14.7% explained by the second and third coordinate respectively. These coordinates (I and II) included all Basmati genotypes except Shaheen Basmati and B.198 while coordinates (III and IV) grouped all Non-Basmati varieties (Figures 5I and 6).
Discussion
Basmati rice from India and Pakistan known as special aromatic rice is circumscribed commercially for gene transformation, and hence, has been maintained with little variability across populations. Due to high value of Basmati rice in foreign exchange, its adulteration is a common issue. Moreover, Varietal homozygosity and lack of inter-population variation among Basmati varieties reinforce the determination of its genetic structure and identification of variety specific marker. Bulk segregant analysis (BSA) is a fast and inexpensive approach that avoids the necessity to genotype population on individual basis. The technique also allows probes candidate genes for a trait to be tested for differences in allele frequency between lines selected to differ specifically in that trait. In past few years, several analyses have been performed using RAPD and ISSR-BSA to solve genome wide relationships among different species. RAPDs have successfully been used for cultivar analysis in a number of plant species including walnut [16], wheat [17], lolium [18], and stevia [19] for being technically simpler than other DNA based markers. The advantage in the use of ISSR marker lies in their linkage to SSR loci which are likely to mark gene rich regions [20]. The variety specific markers using 10-mer arbitrary primers have been successfully used in distinguishing the rice varieties of distinctly different origins and with distinctly different characteristics.
In our study, a considerable level of variability was observed among ten different cultivars, though, in most of the cases, basmati and other aromatic cultivars exhibited similar banding patterns in BSA. RAPD markers exhibited several bands that were shared among the basmati and other non-basmati fine cultivars, whereas few primers generated the distinctive bands that were not amplified in other tested genotypes and were considered to be variety specific alleles. Allele number and polymorphism for each of the 29 evaluated RAPDs primer differed significantly in its ability to determine variability among the cultivars and ranged from 17 to 2 alleles per marker with an average of 12.4 alleles per locus. This observed pattern was reliable with the report based on much larger RAPD markers with same sample size [21]. Identified level of polymorphism with 262 random amplified polymorphic alleles was ranged from 25-94.4% depending on primers used (Table 2). Similar levels of polymorphism have been reported previously among various panels of rice genotypes under different analysis [22]). It has been reported that the ability to resolve genetic variation may be more directly related to the number of polymorphisms detected by the marker system [23]. One of the reasons for this high level of polymorphism could be that the intra-specific variation in rice is extensive [24].
Previously various studies has already been done with RAPD and ISSR markers for variety specific allele mining and genetic diversity estimation identification such as in cardamom [25] and this marker has proved its efficiency. Here ISSR analysis was an efficient tool for diversity analysis, and differentiation of rice landraces on the basis of polymorphism. Basmati landraces except B. 198 (Figure 3 Cluster I) were grouped together showing similarity at genomic level and difference from other landraces. The reason for B.198 out grouping by ISSR marker might be the diverse genetic background because it meets with Basmati-Pak at o.56. B.198 is also dissimilar with Non-Basmati varieties Supri (7.7) and Pak-386 (7.1%). Thus, these two groups are distinctly diverse (4.85%). The intraspecific polymorphism (75.02%) observed by ISSR markers is higher than interspecific piolymorphism (24.98%) (Figure 5III).
High PIC values can be attributed to the use of more informative markers [26]. It was also found that the higher the PIC value of a locus, the higher the number of alleles detected [27]. The PIC values indicated by GLA-17 (0.89) and UBC-810 (0.89) might be the best marker for diversity analysis of rice varieties while UBC-834 (0.26) and GLN-15 (0.314) are least polymorphic primers (Table 2). Thus, these loci may be useful tools in future genetic studies of rice germplasm.
An allele that was observed in only one or two of the 10 cultivars was considered rare. Our study identified 3 rare alleles of sizes, 650 bp in variety Pk-386 with primer OPN-05, 1075 bp in Super Fine and 800 bp in each Supra and Pak-386 with OPE-09 (Figure 1). Among ISSR, UBC- 814 produces allele of approximately 350 bp in Supri and Basmati. 386. This marker also amplifies a rare allele in Super. Fine of approximately 1500 bp in size. Rare allele for Supri was detected by UBC-820 with band size of 950 bp while in case of B. Pak and S. Basmati, rare allele of sizes 1400 bp and 250 bp was observed in UBC811 and UBC 808 respectively. Moreover, UBC-821 amplifies Super Fine and Basmati. 385 specific allele of size 720 bp that was absent in other varieties. The presence of rare alleles in these cultivars indicates that the materials could be used to estimate the adulteration in basmati varieties and useful for plant breeders and geneticists for future breeding programs as well.
Values of genetic diversity in terms of similarity matrix explained the probability of two randomly chosen alleles originated from different ancestors. In our study, Pair-wise genetic similarity coefficients with RAPD marker varied from 0.527 to 0.849 with an average similarity of 0.67 (Table 3 lower diagonal). This could be the result of reduced intraspecific variations in Pakistani rice due to the common ancestors and the selection of primers for few selected traits. Moreover, these findings are in consistent with our previous findings among 19 rice genotypes of Pakistan using 40 RAPDs with genetic similarity values ranged from 0.4-0.85 [28]. Another study on 42 elite rice varieties which have been commonly used in Indian breeding programs also reported narrow genetic variability in improved cultivars and similarity values ranged from 0.59 to 0.95 [29].
Cluster analysis based on jaccards similarity co-efficient [14] divide the 10 rice cultivars into two major clusters, which effectively differentiated the basmati (Basmati-Shaheen, Basmati-385, Basmati- Pak, Basmati-2000, B-198 and Super Basmati) and other quality rice cultivars from the long non-aromatic coarse cultivars (Supri, Super fine, Supra and Pak-386). These findings are consistent with the studies conducted by some others reports [30,31]. Supri and Super fine in cluster II were found 76.5% similar while Pk-386 met both of these varieties at 0.708 genetic similarity coefficients. Moreover, landrace supra were found 69.6% similar to these varieties. A higher genetic similarity co-efficient of 68% among Basmati cultivars in cluster I revealed low genetic variability and conserveness of alleles. B-Shaheen and B-385 were very close with similarity co-efficient of 85% while B-2000 showed 82% (0.822) similarity with B-198. Moreover, Super Basmati variety with RAPD markers was found distinct from other aromatic varieties in Basmati group and met them at genetic similarity estimates of 0.685 (68%). Similarly, B.198 with ISSR markers showed outcrossing. The most plausible explanation of this is, it might be due to very specific characteristics present in Super Basmati and B.198 but absent in remaining aromatic cultivars. Dendrogram also revealed that cultivars in the same group usually shared a high proportion of ancestry and/or agronomic characteristics such as height, maturity, quality traits etc. Overall genetic diversity among aromatic varieties was quite low (32%). This was expected because of evidences of close association of morphological characters due to selection for similar characteristics [22] and their origin from similar parent B-370.
The dendrograms generated by RAPD and ISSR markers grouped the cultivars differently. This can be explained by two factors: differences in the number of markers (29 and 18 by RAPD and ISSR, respectively) generated by the two techniques, and the fact that ISSR primers target-specific genome regions, whereas RAPD primers amplify arbitrary regions [32]. For instance, the UPGMA cluster analysis of the cultivars based on combined RAPD and ISSR data illustrated that the dendrogram at a similarity of 0.65, resulted in two clusters, with five Basmati cultivars falling to main cluster I and Non-Basmati to cluster II (Figure 4). In addition, the grouping obtained from PCoA confirmed that obtained from UPGMA dendogram.
Our results demonstrated the ability of BSA along with RAPD and ISSR markers to provide a baseline for variety specific marker development. The adaptation of these identified rare alleles in Sequence Characterized Amplified Regions (SCARs) and sequence tagged sites (STS) will be useful for checking Basmati adulteration and effective in rice breeding programs by preserving the integrity of basmati rice. This will also provide the baseline for future management, planning and research.
References
- Akram M (2009) Aromatic rices of Pakistan-a review. Pak J Agric Res 22: 3-4.
- Sakamoto T, Matsuoka M (2008) Identifying and exploiting grain yield genes in rice. CurrOpin Plant Biol 11: 209-214.
- Ahmad MA, Gaur R, Gupta M (2012) Comparative biochemical and RAPD analysis in two varieties of rice (Oryza sativa) under arsenic stress by using various biomarkers. J Hazard Mater 30: 141-148.
- Vikram P, Swamy BPM, Dixit S, Ahmed HU, Sta Cruz MT, et al. (2012) Bulk segregant analysis: “An effective approach for mapping consistent-effect drought grain yield QTLs in rice”. Field Crops Research 134:185-192.
- Venuprasad R, Bool ME, Quiatchon L, Atlin GN (2012) A QTL for rice grain yield in aerobic environments with large effects in three genetic backgrounds. TheorAppl Genet 124: 323-332.
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18: 6531-6535.
- Okun DO, Kenya EU, Oballa PO, Odee DW, Muluvi GM (2008) Analysis of genetic diversity in Eucalyptus grandis (Hill ex Maiden) seed sources using inter simple sequence repeats (ISSR) molecular markers. Afr J Biotech 7: 2119-2123.
- Penner GA (1996) RAPD analysis of plant genome. In: Jauhar PP (Editor) Methods of genome analysis in plants. CRC Press, Boca Raton, 251–268.
- Esselman EJ, Jianqiamg L, Crawford DJ, Windus JL, Welfe AD (1999) Clonal diversity in the rare Calamagrostis porter spp. Insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA and inter-simple sequence repeat markers. MolEcol 8: 443–451.
- Karp S, Kresovich KV, Bhat WG, Ayad T, Hodgkin (1997) Molecular tools in plant genetic resources conservation: A guide to the technologies. IPGRI Technical Bulletin No. 2. International Plant genetic Resources Institute, Rome Italy.
- Basel S (2011) Efficiency of RAPD and ISSR markers in assessing genetic variation in Arthrocnemummacrostachyum (Chenopodiaceae). Braz arch bioltechnol 54: 859-866.
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19 : 11–15.
- Bornet B, Branchard M (2001) Nonanchored inter-simple sequence repeat (ISSR) markers: reproducible and specific tools for genome fingerprinting. Plant MolBiol Rep 22: 427-432.
- Jaccard P (1908) Nouvellesrecherchessur la distribution flora. Bull Sac Nat 44: 223-270.
- Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314-331.
- Ahmed N, Mir JI, Mir RR, Rather NA, Rashid R, et al. (2012) SSR and RAPD analysis of genetic diversity in walnut (Juglansregia L.) genotypes from Jammu and Kashmir, India. PhysiolMolBiol Plants 18: 149-160.
- Mukhtar MS, Rahman M, Zafar Y (2002) Assessment of genetic diversity among wheat (TriticumaestivumL.) cultivars from a range of localities across Pakistan using random amplified polymorphic DNA (RAPD) analysis. Euphytica 128: 417-425.
- Ma X, Gu XY, Chen TT, Chen SY, Huang LK, et al. (2013) Genetic relationships between Lolium (Poaceae) species revealed by RAPD markers. Genet Mol Res 12: 3246-3255.
- Chester K, Tamboli ET, Parveen R, Ahmad S (2013) Genetic and metabolic diversity in Stevia rebaudiana using RAPD and HPTLC analysis. Pharm Biol 51: 771-777.
- Kojima T, Nagaoka T, Noda K, Oihara (1998) Genetic linkage map of ISSR and RAPD markers in Einkorn wheat in relation to that of RFLP markers. Theoretical and Applied Genetics 96 : 37-45.
- Muthusamy S, Kanagarajan S, Ponnusamy S (2008) Efficiency of RAPD and ISSR markers system in accessing genetic variation of rice bean (Vignaumbellata) landraces. Electronic J Biotech 11: 1-10.
- Pervaiz ZH, Rabbani MA, Shinwari ZK, Masood MS, Malik SA (2010) Assessment of genetic variability in rice (Oryza sativa L) germplasm from Pakistan using RAPD markers. Pak J Bot 42: 3369-3376.
- Sivaprakash KR, Prasanth SR, Mohanty BP, Parid A (2004) A Genetic diversity of black gram landraces as evaluated by AFLP markers. Current Science 86:1411-1415.
- Fugang R, Lu BR, Li S, Haung J, Zhu Y (2003) A comparative study of genetic relationship among the AA-genome using RAPD and SSR markers. TheorAppl Genet 108: 113-120.
- Fugang R, Lu BR, Li S, Haung J, Zhu Y (2003) A comparative study of genetic relationship among the AA-genome using RAPD and SSR markers. TheorAppl Genet 108: 113-120.
- Akkaya MS, Buyukunal-Bal EB (2004) Assessment of genetic variation of bread wheat varieties using microsatellite markers. Euphytica 135:179-185.
- Wong SC, Yiu PH, Bong STW, Lee HH, Neoh PNP et al., (2009) Analysis of Sarawak Bario rice diversity using microsatellite markers. Am J AgricBiolSci 4: 298-304.
- Arif M, Kousar S, Bajwa MA, Arif A, Zafar Y (2005) Genetic diversity among rice genotypes of Pakistan through random amplified polymorphic DNA analysis. Pak J Bot 37: 585-592.
- Davierwal AP, Chowdari KV, Kumar S, Reddy APK, Ranjekar PK, et al. (2000) Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica 108: 269-284.
- Nagaraju J, Kathirvel M, Kumar RR, Siddiq EA, Hasnain SE (2002) Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. ProcNatlAcadSci U S A 99: 5836-5841.
- Rabbani MA, Pervaiz ZH, Masood MS (2008) Genetic diversity analysis of traditional and improved cultivars of Pakistani rice (Oryza sativa L) using RAPD markers. Elect J Biotechnol 11: 1-10.
- Martins-Lopes P, Lima-Brito J, Gomes S, Meirinhos J, Santos L, et al. (2007) RAPD and ISSR molecular markers in Oleaeuropaea L.: genetic variability and cultivar identification. Genet Resour Crop Evol 54:117–128.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 434232
- [From(publication date):
August-2016 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 433246
- PDF downloads : 986