Research Article Open Access
Use of Agro Industrial Residues for the Production of Amylase by Penicillium sp. for Applications in Food Industry
Nidhi Arora*, Supreet Kaur and Sawinder KaurDepartment of Biotechnology, Lovely Professional University, Phagwara, Punjab, India
- *Corresponding Author:
- Arora N
Department of Biotechnology
Lovely Professional University
Phagwara, Punjab, India
Tel: +9666665287
E-mail: nidhi.arora2492@gmail.com
Received date: April 13, 2017; Accepted date: April 28, 2017; Published date: May 05, 2017
Citation: Arora N, Kaur S, Kaur S (2017) Production of Amylase by Penicillium sp. for Applications in Food Industry. J Biotechnol Biomater 7:256. doi:10.4172/2155-952X.1000256
Copyright: © 2017 Arora N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Glucoamylase production has been investigated by solid state fermentation using agro-industrial residues, rice bran, wheat bran and banana peel by Penicillium sp. Out of the different agricultural residues tested for enzyme production, rice bran yielded the maximum glucoamylase activity (10.29 ± 0.07 Units/ml), protein content (1.22 ± 0.0014 mg/ml) and specific activity (8.4 Units/mg protein) with Penicillium sp. after 7th day of fermentation as compared to other agro residues. Media supplementation with carbon and nitrogen sources enhances the enzyme activity. For maximum production of enzyme, sucrose (13.70 ± 0.77 Units/ml) (1% mass level) and yeast extract (14.41 ± 0.07Units/ml) (1% mass level) as carbon and nitrogen source respectively were found optimum on rice bran with Penicillium sp. Optimum enzyme activity was observed at 90°C, pH 9. The partial purification of enzyme from rice bran by Penicillium sp. was done with 60% ammonium sulphate precipitation showed maximum enzyme activity. Enzyme hydrolysis showed maximum activity (3.69 Units/ml) with potato starch from rice bran by Penicillium sp.
Keywords
Amylase; Glucoamylase; Asperigillus; Rhizopus; Bradford method; Penicillium sp.; Enzyme
Introduction
Recent discoveries on the use of microorganisms as sources of industrially relevant enzymes have led to an increased in the application of microbial enzymes in various industrial processes. Nowadays, the use of microorganism as biotechnological source of industrially relevant enzymes has interest in exploration of extracellular enzymatic activities in several microorganisms. Amylases are used in dextrose production, in the baking industry, in the brewing of low-calorie beer and in whole gain hydrolysis for the alcohol industry [1].
The amylase family has two major classes, namely amylase (EC 3.2.1.1) and glucoamylase (EC 3.2.1.3). Alpha-amylase are extracellular enzymes which hydrolyze starch into maltose, glucose and maltotriose by cleaving the 1,4-D- glucosidic linkages between adjacent glucose units in the linear amylose chain [2].
Glucoamylase (GA)
Hydrolyses single glucose units from the non-reducing ends of amylase and amylopectin in a stepwise manner and produce glucose as the sole end-product from and related polymers. Unlike alpha amylase, most glucoamylases are also able to hydrolyse the 1,6-linkages at the branching points of amylopectin, at a lower rate than 1,4-linkages [3]. Alpha amylase is secreted as a primary metabolite and its secretion is growth associated [4]. It is an important group of enzymes in starch processing. They are second to the proteases in worldwide distribution and sales among industrial enzymes.
Production of glucoamylase (GA) is performed mostly by two methods, Solid State or Solid-Substrate Fermentation (SSF) and Submerged Fermentation (SMF) [5]. SSF has many advantages over SMF, such as simple technique, superior productivity, low capital investment, low energy requirement and less water output, better product recovery and lack of foam build up and reported to be the most appropriate process for developing countries [6]. Selection of a suitable microorganism is one of the most important in SSF. Several groups of microorganism such as filamentous fungi group can grow on solid substrates.
Agro-industrial residues are generally considered the best substrates for the SSF processes and enzyme production by SSF is not an exception to that [7]. Use of suitable low cost fermentation medium for production of α-amylase by using agricultural by products has been reported [8].
Fungal species such as Asperigillus, Rhizopus and Penicillium are considered to be the chief producers of amylase [9,10]. Among large number of agro-industrial wastes (wheat bran, wheat straw, rice bran, rice husk, rice straw, sago hampas, grapevine trimmings dust, saw dust, corncobs, coconut coir pith, banana waste, tea waste, cassava waste, palm oil mill waste, aspen pulp, sugar beet pulp, sweet sorghum pulp, apple pomace, peanut meal, rapeseed cake, coconut oil cake, mustard oil cake, cassava flour, wheat flour, corn flour, steamed rice, steam pretreated willow, starch and bagasse ) [11] are available as substrate for the production of enzyme.
In the present study glucoamylase production has been studied with a natural isolate of the fungus Penicillium sp. using the rice bran waste obtained from local market of Haryana along with wheat bran and banana peel as the growth substrates.
Methods
Microorganism and its maintenance
Penicillium sp. was propagated on Potato Dextrose Agar (PDA) medium. Slants were grown at 30°C for 5 days and stored at 4°C. Subculturing was done after every 15 days.
Preparation of basal media
Solid state fermentation with the agro residues wheat bran (WB), Rice Bran (RB) and Banana Peel (BP) was done by moistening with mineral salt solution having composition (mg/gds) (NH4)2SO4 4, MgSO4.7H2O 1, FeSO4.7H2O 0.02, K2HPO4 1.4 and KH2PO4 0.6.The pH of the media was adjusted to 7.0 [5].
Enzyme production and extraction
The production media contained 10 g of solid substrate and 20 ml mineral solution in 250 ml Erlenmeyer flasks initially maintained at pH 7. The media were inoculated with 2 ml of inoculum having 106 spores/ ml collected from 72 h grown culture of Penicillium sp. at 28 ± 2°C. Inoculated production media were incubated under static conditions at 28 ± 2°C and enzyme production was checked after every 24 h for 5 days. Enzyme was extracted in 100 ml, 0.1 M sodium acetate buffer on a rotary shaker at 100 rpm for 60 min. The content was filtered through muslin cloth and the filtrate was used as the enzyme source.
Measurement of protein content and enzyme activity of crude enzyme extract
Protein estimation: Protein content was determined by using Bradford method. Bradford reagent was made by dissolving 100 mg Coomassie Blue G-250 in 50 ml of 85% ethanol and adding 100 ml of 85% (w/v) phosphoric acid and make volume to 1 L. 1 ml of Bradford reagent was added to protein sample and incubated at room temperature for 10 min. Standard curve was prepared by using BSA, stock solution having concentration of 100 mg/ml. The absorbance of each sample was measured at 595 nm using a UV-visible spectrophotometer [12].
Enzyme activity: The enzyme activity was determined by incubating a reaction mixture containing 0.95 ml of 0.1 M sodium acetate buffer (pH 5), 1.0 ml starch solution (1%, w/v) and 0.05 ml of crude enzyme at 55°C for 15 min [13]. The reducing sugar released after 10 min were measured with 3,5-dinitrosalicyclic acid (DNSA) reagent [14] using glucose as a standard. Glucoamylase activity unit (U) was expressed as the amount of enzyme releasing 1 μmol of glucose equivalent per min under assay condition and enzyme activity is expressed in terms of units per gram dry fermented substrate (U/gdfs).
Growth kinetics
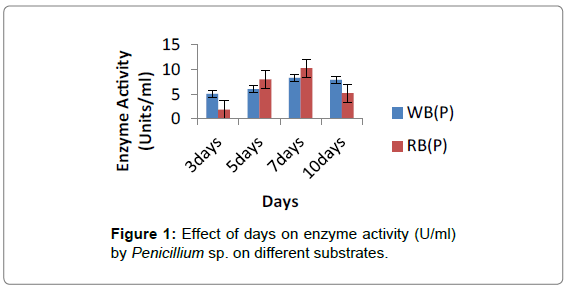
Wheat bran, rice bran and banana peel (10 g) were kept separately in a 250 ml flask and then moistened with 20 ml of mineral solution containing (mg/gds) (NH4)2SO4 4, MgSO4.7H2O 1, FeSO4.7H2O 0.02, K2HPO4 1.4 and KH2PO4 0.6, initially maintained at pH 7 [5] and sterilized at 121°C for 30 min. The fermentation process was started by adding 2 ml of spore suspension as prepared above. The whole content was mixed thoroughly and then incubated at 30°C for 3 days, 5 days, 7 days and 10 days in a stationary condition.
Separation and identification of sugars by thin layer chromatography
Sugars get separated on the basis of differential adsorption onto silica gel. The reaction mixture contains 1 ml of enzyme and 1 ml of standard solution which was prepared as 1% solution of standard sugars such as glucose, maltose in 10% isopropanol (v/v).Carefully the solution of individual standard sugars and the mixture of the samples applied on the separate marked spots. The chromatogram was developed with solvent system of ethyl acetate:iso-propanol:water:pyridine (26:14:7:2, v/v). Due to capillary action solvent reaches the marked line on the upper side of the silica plate. Plate was removed from chromatographic chamber and let it dry at room temperature. The spots were visualised by spraying aniline diphenylamine reagent (mix 5 volumes of 1% aniline and 5 volumes of 1% diphenylamine in acetone with 1 volume of 85% phosphoric acid) and heating at 100°C. The sugar spots appeared as dark brown spots.
Identification of the sugars was done by comparing the relative fraction (Rf) values of the samples with that of the standards.
Rf=Distance travelled by unknown amino acid/Distance travelled by solvent system
Characterization of glucoamylase
Soluble starch used in assay system for enzyme-substrate reaction was supplemented as 1%. Enzyme activity was checked individually at four different pH as 3, 5, 7 and 9 of the 0.1 M Sodium acetate buffer used and at four different temperatures 35°C, 55°C, 75°C and 90°C.
Effect of temperature: To check the optimum temperature for glucoamylase activity, crude enzyme of 7th day extract was incubated with 1% soluble starch in 0.1 M acetate buffer, pH 5 at different temperatures as 35°C, 55°C, 75°C and 90°C. Enzyme activity was checked individually.
Effect of pH: To check the optimum pH for glucoamylase activity, the pH of 0.1 M sodium acetate buffer was varied as 3, 5, 7 and 9 and the enzyme activity was checked individually.
Effect of carbon and nitrogen additives: The optimum SSF medium of wheat bran and rice bran was supplemented with different carbon (glucose, fructose, sucrose and starch) and nitrogen sources (urea, yeast extract, soybean meal and peptone) at 1% (by mass) level to study their effect on GA production by Penicillium sp. The fermentation was carried out at pH 5 and 30°C for 7 days [15].
Partial purification of crude enzyme
Ammonium sulfate precipitation: 100 ml of the crude enzyme were first brought to 40% saturation with solid ammonium sulfate (enzyme grade) and the process was carried out at 4°C [16]. The whole apparatus was placed on a magnetic stirrer and ammonium sulphate was allowed to dissolve in the enzyme solution. The precipitated proteins were regimented by centrifugation for 15 min at 1000 rpm. The resulted pellet was dissolved in 3 ml of 0.2 M phosphate buffer at pH 6.2.The supernatant was applied again with ammonium sulfate to achieve 60% saturation. Both enzyme activity and protein content were determined for each separate fraction [17].
Dialysis: Pellet was dissolved in sodium acetate buffer (0.2 M) and dialysed against the same buffer (pH 5) with 3 equal changes of buffer after every hour to ensure the complete removal of the ammonium salt, enzyme was dialysed for 3 h at 4°C. The dialysed fraction referred to as partially purified glucoamylase [18].
Application of glucoamylase
Enzyme hydrolysis of various starches: The ability of the crude amylase to hydrolyze different native starches was studied using corn and potato starches. Commercially available soluble starch was used as standard. The assay mixture consisted of 1% of various starch in 0.2 M citrate-phosphate buffer (pH 6) and 0.05 of the enzyme. Enzyme assay was carried out after incubation at 40°C for 10 min [19].
Results and Discussion
Glocoamylase production using solid substrates
Rice bran, wheat bran and banana peel was used as substrate for the production of glucoamylase by Penicillium sp. for 5 days in solid state fermentation. The results showed that maximum activity was shown by Penicillium sp. on rice bran (8.029 ± 0.014 Units/ml) followed by wheat bran and banana peel.
Growth kinetics
Wheat Bran, Rice Bran and Banana Peel (10 g) which were incubated for 3 days, 5 days, 7 days and 10 days in stationary condition showed enzyme activity. Penicillium sp. on rice bran (10.29 ± 0.07 Units/ml) at 7th day showed maximum enzyme activity followed by wheat bran and banana peel. After 7 days enzyme activity decreased (Figure 1).
Thin layer chromatography of hydrolysis products
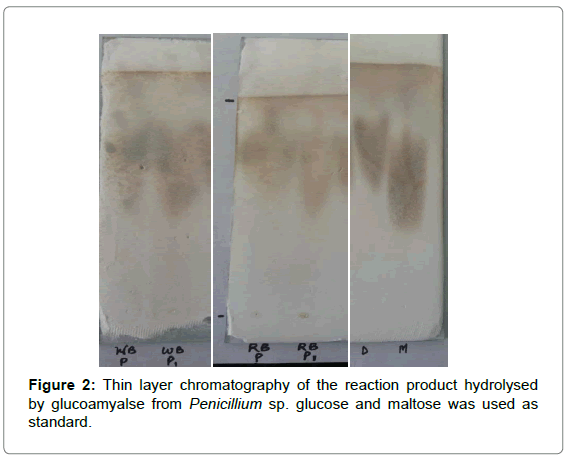
Glucoamylase is an exo-acting enzyme that catalyses the production of β-glucose from the non-reducing ends of amylase, amylopectin and glycogen. It consecutively hydrolyses α-1,4; α-1,6 and rare α-1,3 linkages [20]. TLC revealed only glucose as the hydrolysis product of starch which indicates that the crude enzyme was a glucoamylase [21]. Rf values of WB(A),WB(P), RB(A) and RB(P) are 0.85, 0.78, 0.75 and 0.89, respectively and Rf values of glucose and maltose of WB(A), WB(P), RB(A) and RB(P) are 0.85,0.89; 0.78,0.74; 0.75,0.7; 0.89,0.85, respectively (Figure 2).
| Substrates | Enzyme Activity(Units/ml) | |
|---|---|---|
| 40% | 60% | |
| WB (A) | 9.641 ± 0.037 | 9.84 ± 0.044 |
| WB (P) | 14.52 ± 0.014 | 8.62 ± 0.035 |
| RB (A) | 14.24 ± 0.014 | 6.15 ± 0.037 |
| RB (P) | 13.86 ± 0.022 | 14.32 ± 0.014 |
Table 1: Enzyme activity (Units/ml) of partial purified enzyme.
Characterization of enzyme
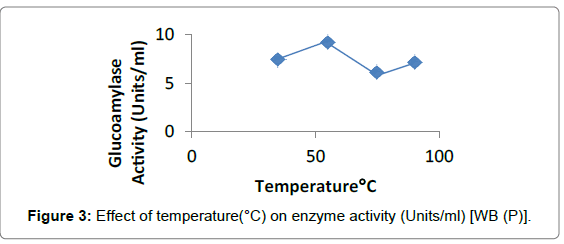
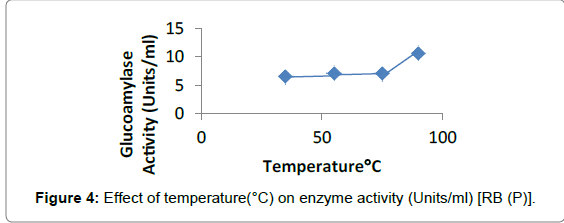
Effect of temperature: Glucoamylase activity was tested at different temperatures. Penicillium sp. showed the maximum activity in case of WB (9.23 ± 0.10 Units/ml) at 55°C and for RB (10.66 ± 0.0007 Units/ml) at 90°C (Figures 3 and 4).
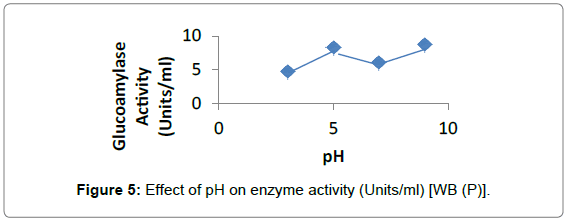
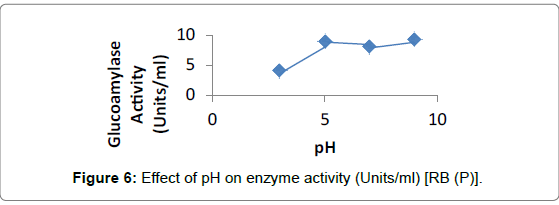
Effect of pH: Enzyme activity was maximum at pH 9, for Penicillium sp. on RB and WB using 1% soluble starch for enzyme assay (Figures 5 and 6).
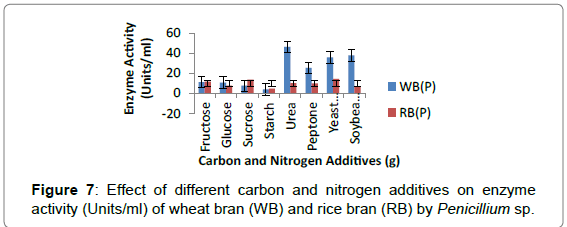
Effect of carbon sources and nitrogen additives on enzyme activity: Addition of different sugars as additives to wheat bran and rice bran medium enhanced glucoamylase production by Penicillium sp. The maximum activity was shown by Penicillium sp. on RB with sucrose (13.70 Units/ml) as carbon source and yeast extract (14.41 Units/ml) as nitrogen source (Figure 7).
Partial purification of glucoamylase
Glucoamylase was purified by ammonium sulfate precipitation and dialysis process. Penicillium sp. in rice bran and wheat bran showed an increase in enzyme activity as comparative to crude enzyme (Table 1).
Application of glucoamylase
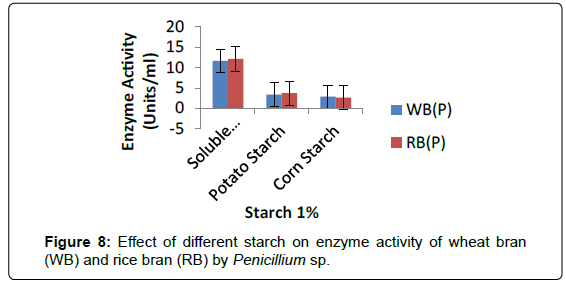
Enzyme hydrolysis of various starches: The rate of the amylase hydrolysis of different native starches was evaluated. Figure 8 showed that the amylase from the Penicillium sp. and A. terreus was able to hydrolyse different starches to varying degrees. Highest rate of hydrolysis was observed for potato starch, followed by corn starch (Figure 8).
Conclusion
On the basis of the finding of the present study it can be concluded that out of the all different agricultural residues which is used to test for enzyme production, rice bran has the maximum glucoamylase activity (10.29 ± 0.07 Units/ml), protein content (1.22 ± 0.0014 mg/ml) and specific activity (8.4 Units/mg protein) with Penicillium sp. at 7th day as compared to other agro residues. By usage of Media supplements with carbon and nitrogen sources increases the enzyme activity. Sucrose (13.70 ± 0.77 Units/ml) (1% mass level) and yeast extract (14.41 ± 0.07 Units/ml) (1% mass level) as carbon and nitrogen source respectively were found optimum on rice bran with Penicillium sp. Optimum enzyme activity was observed at 90°C, pH 9.The partial purification of enzyme from rice bran by Penicillium sp. was done using ammonium precipitation and showed maximum enzyme activity at 60%. Enzyme hydrolysis showed maximum activity (3.69 Units/ml) with potato starch from rice bran by Penicillium sp.
References
- Duncan R (2003) The dawning era of polymer therapeutics. Nat Rev Drug Discov 2: 347-360.
- Ferrari M (2005) Cancer nanotechnology: Opportunities and challenges. Nat Rev Cancer 5: 161-171.
- Lin Y, Taylor S, Li H, Shiral Fernando K, Qu L, et al. (2004) Advances toward bioapplications of carbon nanotubes. J Mater Chem 14: 527âÂ?Â?541.
- De Jong WH, Borm PJ (2008) Drug delivery and nanoparticles: Applications and hazards. Int J Nanomedicine 3: 133-149.
- Pantarotto D, Partidos CD, Graff R, Hoebeke J, Briand JP, et al. (2003) Synthesis, structural characterization and immunological properties of carbon nanotubes functionalized with peptides. J Am Chem Soc 125: 6160.
- Pantarotto D, Partidos CD, Hoebeke J, Brown F, Kramer E, et al. (2003) Immunization with peptide-functionalized carbon nanotubes enhances virus-specific neutralizing antibody responses. J Am Chem Soc 10: 961-966.
- Vardharajula S, Ali SZ, Tiwari PM, EroÃÂ?Ÿlu E, Vig K, et al. (2012) Functionalized carbon nanotubes: biomedical applications. Int J Nanomedicine 7: 5361-5374.
- Pantarotto D, Briand JP, Prato M, Bianco A (2004) Translocation of bioactive peptides across cell membranes by carbon nanotubes. Chem Commun 1: 16-17.
- Madani SY, Naderi N, Dissanayake O, Tan A, Seifalian AM (2011) A new era of cancer treatment: Carbon nanotubes as drug delivery tools. Int J Nanomedicine 6: 2963-2979.
- Al-Jamal KT, Kostarelos K (1996) Science of fullerenes and carbon nanotubes: Imaging carbon nanotubes in vivo: A vignette of imaging modalities at the nanoscale. Pan Stanford Publishing, Singapore.
- Pastorin G, Kostarelos K, Prato M, Bianco A (2005) Functionalized carbon nanotubes: Towards the delivery of therapeutic molecules. J Biomed Nanotechnol1: 1-10.
- Dresselhaus MS, Dresselhaus G, Eklund PC (1996) Science of fullerenes and carbon nanotubes. CA: Academic Press, San Diego.
- Mahmood M, Karmakar A, Fejleh A, Mocan T, Iancu C, et al. (2009) Synergistic enhancement of cancer therapy using a combination of carbon nanotubes and anti-tumor drug. Nanomedicin 4: 883-893.
- Balavoine F, Schultz P, Richard C, Mallouh V, Ebbesen T (1999) Helical crystallization of proteins on carbon nanotubes: A first step towards the development of new biosensors. Angewandte Chemie International Edition 38: 1912-1915.
- Tsang SC, Davis JJ, Malcolm L, Green H, Allen H, et al. (1995) Immobilization of small proteins in carbon nanotubes: High-resolution transmission electron microscopy study and catalytic activity. J Chem Soc 17: 1803-1804.
- Tsang SC, Guo Z, Chen YK, Green MLH, Allen H, et al. (1997) Immobilization of platinated and iodinated oligonucleotides on carbon nanotubes. Angewandte Chemie International Edition 36: 2197-2198.
- Ganji MD (2010) Calculations of encapsulation of amino acids inside the (13, 0) single-walled carbon nanotube. Fullerenes, Nanotubes and Carbon Nanostructures 18: 24-36.
- Dawson RMC (1959) Data for biochemical research. Clarendon Press, Oxford.
- Doolittle RF (1989) Redundancies in protein sequences. Plenum, New York.
- Davies JS (2006) Amino acids, peptides and proteins. R Soc Chem.
- Kendall EC, McKenzie BF (1929) A publication of reliable methods for the preparation of organic compounds. Org Synth 9: 4.
- David LN, Michael MC (2005) Principles of biochemistry. WH Freeman, New York.
- Koski RR (2008) Omega-3-acid ethyl esters (lovaza) for severe hypertriglyceridemia. P T 33: 271-303.
- Paul B, Watkins MD, Neil K, John T, Connie R, et al. (2006) Amino transferase elevations in healthy adults receiving 4 grams of acetaminophen daily a randomized controlled trial. J Am Med Assoc 296: 87-93.
- Huajian G, Yong K, Daxiang C (2003) Spontaneous insertion of DNA oligonucleotides into carbon nanotubes. Nano Lett 3: 471-473.
- Yang W, Thordarson P, Gooding JJ, Ringer SP, Braet F (2007) Carbon nanotubes for biological and biomedical applications. Nanotechnology 18: 4951-4957.
- Balasubramanian K1, Burghard M (2006) Biosensors based on carbon nanotubes. Anal Bioanal Chem 385: 452-468.
- Salimi A, Compton RG, Hallaj R (2004) Glucose biosensor prepared by glucose oxidase encapsulated sol-gel and carbon-nanotube-modified basal plane pyrolytic graphite electrode. Anal Biochem 333: 49-56.
- Salvador-Morales C, Flahaut E, Sim E, Sloan J, Green ML, et al. (2006) Complement activation and protein adsorption by carbon nanotubes. Mol Immunol 43: 193-201.
- Lu AJ, Pan BC (2005) Interaction of hydrogen with vacancies in a (12, 0) carbon nanotube. Phys Rev B 71: 165416.
- Liu GR, Cheng Y, Dong M, Li ZR (2005) A study on self-insertion of peptides into single-walled carbon nanotubes based on molecular dynamics simulation. Int J Mod Phys C 16: 1239-1250.
- Cheng Y, Liu GR, Li ZR, Liu C (2006) Computational analysis of binding free energies between peptides and single-walled carbon nanotubes. Physica A 367: 293-403.
- Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NWS, Shim M, et al. (2003) Non-covalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proc Natl Acad Sci 100: 4984-4989.
- Besteman K, Lee J, Wiertz FGM, Heering H, Dekker C (2003) Enzyme-coated carbon nanotubes as single-molecule biosensors. Nano Lett 3: 727-730.
- Star A, Gabriel JCP, Bradley K, Gruner G (2003) Electronic detection of specific protein binding using nanotube FET devices. Nano Lett 3: 459-463.
- Trzaskowski B, Jalbout AF, Adamowicz L (2006) Molecular dynamics studies of protein fragment models encapsulated into carbon nanotubes. Chem Phys Lett 430: 97-100.
- Roman T, Dino WA, Nakanishi H, Kasai H (20006) Amino acid adsorption on single walled carbon nanotubes. Eur Phys 38: 117-120.
- Vardanega D, Picaud F (2009) Detection of amino acids encapsulation and adsorption with dielectric carbon nanotube. J Biotechnol 144: 96-101.
- Chang CM, Tseng HL, Leon AD, Jalbout AF (2013) Theoretical study of amino acids encapsulation in zig-zag single-walled carbon nanotubes. J Comput Theor Nanosci 10: 521-526.
- Zhao J, Buldum A, Han J, Lu JP (2002) Gas molecule adsorption in carbon nanotubes and nanotube bundles. Nanotechnology 13: 195-200.
- Ganji MD (2008) Theoretical study of the adsorption of CO2 on tungsten carbide nanotubes. Phys Lett 372: 3277.
- Pupysheva OV1, Farajian AA, Yakobson BI (2008) Fullerene nanocage capacity for hydrogen storage. Nano Lett 8: 767-774.
- Ganji MD (2008) Behavior of a single nitrogen molecule on the pentagon at a carbon nanotube tip: A first-principles study. Nanotechnology 19: 025709-025714
- Contreras-Torres FF, Jalbout AF, Jimenez-Fabian I, Amelines OF, Basiuk V (2008) Interaction of cation-encapsulated single-walled carbon nanotubes with small polar molecules. J Phys Chem 112: 2736-2742.
- Cottrell LT (1958) The strengths of chemical bonds. Butterworths Scientific Publications, London.
- Hirschfelder JO, Curtiss CF, Byron RB (1964) Molecular theory of gases and liquids. Society for Industrial and Applied Mathematics, University of Winsconsin, Madison.
- Cox BJ, Thamwattana N, Hill JM (2006) Mechanics of atoms and fullerenes in single walled carbon nanotubes. Proc R Soc 463: 461-476.
- Al Garalleh H, Thamwattana N, Cox BJ, Hill JM (2016) Encapsulation of L-Histidine amino acid inside single walled carbon nanotubes. J Biomater Tissue Eng 6: 362-369.
- Cox BJ, Thamwattana N, Hill JM (2008) Mechanics of nanotubes oscillating in carbon nanotube bundles. Proc R Soc 464: 691-710.
- Rappi AK, Casewit CJ, Colwell KS, Goddard WA, Skid WM (1992) UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J Am Chem Soc 114: 10024-10035.
- Anslyn E, Dougherty DA (2006) Modern physical organic chemistry. University Science Books.
- Benson SW (1965) III-bond energies. J Chem Educ 42: 502.
- Darwent B (1970) National standard reference data series. National Bureau of Standards, Washington DC.
- Dresselhaus MS, Dresselhaus G, Satio R (1995) Physics of carbon nanotubes. Carbon 33: 883âÂ?Â?891.
- Cox B, Thamwattana N, Hill JM (2006) Mechanics of atoms and fullerenes in single walled carbon nanotubes. Proc R Soc 463: 461-476
- Wilder JWG, Venema LC, Rinzler AG, Smalley RE, Dekker C (1998) Electronic structure of atomically resolved carbon nanotubes. Nature 391: 59-62.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 5222
- [From(publication date):
June-2017 - Jul 05, 2025] - Breakdown by view type
- HTML page views : 3933
- PDF downloads : 1289