Research Article Open Access
UPLC-ESI-Q-TOF-MS/MS Characterization of Phenolics from Crataegus monogyna and Crataegus laevigata (Hawthorn) Leaves, Fruits and their Herbal Derived Drops (Crataegutt Tropfen)
Mohamed Gamaleldin Elsadig Karar and Nikolai Kuhnert*
Department of Life Sciences and Chemistry, Jacobs University Bremen, Campus Ring 8, 28759 Bremen, Germany
- *Corresponding Author:
- Nikolai Kuhnert
Department of Life Science and Chemistry
Jacobs University Bremen, Campus Ring 8, 28759 Bremen, Germany
Tel: 49 421 200 3120
Fax: 49 421 200 3229
E-mail: n.kuhnert@jacobs-university.de
Received date: September 17, 2015; Accepted date: November 03, 2015; Published date: November 10, 2015
Citation: Karar MGE, Kuhnert N (2015) UPLC-ESI-Q-TOF-MS/MS Characterization of Phenolics from Crataegus monogyna and Crataegus laevigata (Hawthorn) Leaves, Fruits and their Herbal Derived Drops (Crataegutt Tropfen). J Chem Biol Ther 1:102. doi: 10.4172/2572-0406.1000102
Copyright: © 2015 Karar MGE, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Chemical Biology & Therapeutics
Abstract
Crataegus species are medicinal plants naturally growing in Europe, Asia and the north of Africa. The plant extracts have been used for a long time in traditional medicine for the treatment of cardiovascular diseases. Many natural health product, including tablets, teas, and aqueous extracts are made from Crataegus species. These products are currently marketed as an alternative therapy for New York Heart Association (NYHA I-III) heart failure. But further studies suggested the use of the plant extracts for various other cardiovascular diseases including hypertension, hyperlipidemia, arrhythmia and angina. Thus, due to the important role that hawthorn plays in medicine and human health, we have investigated qualitatively the phytoconstituents of C. monogyna and C. laevigata, leaves, fruits and their herbal derived drops (Crataegutt Tropfen) using UPLC-ESI-Q-TOF-MS/MS and HPLC-ESI-MSn. A total of 113 compounds were identified, characterized or tentatively assigned on the basis of their accurate mass data generated by Q-TOF-MS, MS/MS, MSn fragmentation patterns, retention behaviors, or by comparison with commercial reference standards and literature data. The identified constituents belonged to chlorogenic acids, phenolic acids, proanthocyanidins, flavonoid glycosides, flavonoid aglycones and derivatives, and other compounds. To our knowledge 63 of the identified phytoconstituents were not reported previously in Crataegus species and two of them for the first time in nature. Additionally, it is important to highlight that this is the first comprehensive study of the bioactive phenolic compounds of C. monogyna and C. laevigata, leaves, fruits and the herbal derived drops (Crataegutt Tropfen).
Keywords
Herbal drugs; natural products; C. monogyna; C. laevigata; polyphenols; flavonoid glycosides; mass spectrometry
Introduction
Crataegus species also known as hawthorn (Rosaceae) are small trees and shrubs naturally growing in Europe, Asia and the north of Africa. Many natural health products, including tablets, teas, and aqueous extracts are made from Crataegus species. Crataegus is described in various pharmacopoeia including German Pharmacopoeia and Chinese Pharmacopoeia. Several reports suggested that hawthorn and the herbal derived drops (Crataegutt Tropfen) could be used as an alternative therapy for various cardiovascular diseases, such as hyperlipidemia, hypertension, arrhythmia, angina and New York Heart Association (NYHA) class I-III heart failure [1,2]. Thus, confirmed the use of hawthorn flowers, leaves and berries, alone or in combination traditionally in Europe for the treatment of a variety of ailments including high blood pressure and heart disorders [3]. However, the Complete German Commission E Monographs declares that Crataegus monogyna and Crataegus laevigata can be used to treat cases of cardiac failure [4].
Polyphenols are natural compounds characterized by a high structural diversity, and many of them are essential components in our diets. They are found only in plants and certain fungal species and are not synthesized by humans or animals [5]. Currently, polyphenols are seen as secondary metabolites characterized by a high range of physiological functions [5]. Several studies have shown that the high consumption of polyphenols have protective effects against cancer and inflammatory diseases [6]. The anti-inflammatory effects of phenolic compounds have been attributed mostly to their antioxidant activity presumably scavengers the reactive oxygen and nitrogen species in vivo [7,8]. However, the antioxidant hypothesis is under intense scrutiny [9]. It is also reported that the phenolics have anti-tumor, antidiabetic, anti-mutagenic and anti-HIV properties [10,11]. Moreover, there are several studies suggesting benefits of polyphenols intake for reducing the risks of cardiovascular problems, skin diseases, asthma, wound healing, protect from drug toxicity and UV radiations [10,12]. Although dietary plants polyphenols have been associated with several beneficial health effects for humans, their bioavailability is still under discussion [13,14].
A number of analytical techniques had been used such as liquid chromatography with diode array detection (HPLC-DAD) and liquid chromatography coupled to electrospray ionization multistage mass spectrometry (LC-ESI-MSn) to identify different classes of phytoconstituents in hawthorn species including proanthocyanidins, phenolic acids, flavonoid glycosides, flavonoid aglycones, triterpene acids, sterols and chlorogenic acids [3,15-18]. Herein, the aim of this study was to improve the knowledge of the methanol: water (2:1) mixture extract of the fruits and leaves of C. monogyna and C. laevigata and the traditionally derived drops (Crataegutt Tropfen) by a comprehensive identification and characterization of their bioactive phenolic compounds by using ultra-performance liquid chromatography coupled to electrospray ionization quadropole-timeof- fight mass spectrometry (UPLC-ESI-Q-TOF-MS) in addition to the LC-ESI-MSn as a powerful analytical techniques. In this contribution, we have tested the methanolic extract of the plant materials in particular, because it known to be more effective in extraction of low molecular weight phenolic compounds. The obtained results may contribute to a better understanding of influence of hawthorn phenolics on biological, nutritional and medicinal prosperities.
Recently, the improvement of ultra-high-pressure pump systems and small size filling substances leads to improve resolution, greater separation, high peak efficiency and reduced solvent consumption and running time compared with usual HPLC. In this regard, the combination of UPLC with MS allows a better separation, identification and characterisation of bioactive compounds in medicinal plants and complex mixtures. Consequently, the mass analyser Q-TOF-MS combines the high efficiency of TOF analysis in both MS and tandem MS (MS/MS) manners, providing high mass accuracy and better sensitivity for both precursor and fragment ions [19,20].
On numerous occasions we have shown that use of modern analytical instrumentation frequently merits reinvestigation of well characterized medicinal or dietary plant material. Using improved resolution and sensitivity on occasions large number of previously overlooked secondary metabolites can be readily identified. For this reasons we decided to re-investigate Crataegus as one of the best and most intensity studied medicinal plants.
Experimental
Chemicals and standards
All the chemicals (analytical grade) and authentic standards of polyphenols were purchased from Sigma-Aldrich, Applichem, HWI analytic, and Phytolab (Germany). 3,4-di-O-caffeoylquinic acid 69, 3,5-di-O-caffeoylquinic acid 70, 4,5-di-O-caffeoylquinic acid 71, epicatechin 30, quercetin 86, quercetin 3-O-(6-O-rhamnosyl-glucoside) (rutin) 65, quercetin 3-O-glucoside 68, kaempferol 7-O-glucoside 63, kaempferol 3-O-rutinoside 59, luteolin 7-O-glucoside 62, luteolin 8-C-glucoside 48, quercetin 3-O-arabinoside, quercetin 3-O-rhamnoside (quercitrin) 77, apigenin 7-O-glucoside 76, phloretin 2’-O-glucoside (phlorizin) 81, p-coumaric acid 44, malic acid 106, ascorbic acid 108 and quinic acid 1 were used as authentic standards.
Plant materials and the herbal drops
Leaves and fruits of C. monogyna and C. laevigata were freshly collected from a garden in Jacobs University, Bremen, Germany. The apple (Malus domestica) fruits were purchased from a local market in Bremen, Germany. The herbal drops (Crataegutt Tropfen, Schwabe, Karlsruhe) was granted from a pharmacy in Bremen, Germany.
Sample preparation
The sample preparation was achieved as previously described [21]. Fresh hawthorn samples (10 g of each) were freeze dried with the liquid nitrogen and crushed by a mortar. Then, the samples were extracted with methanol:water (2:1) mixture by sonication for 30 min and filtered through a Whatman no. 1 filter paper. The solvents were removed by evaporation in vacuo and the extracts were stored at −20°C until required, thawed at room temperature, dissolved in methanol (50 mg/10 mL of methanol), filtered through a membrane filter (0.45 μm) and used directly for UPLC-ESI-Q-TOF-MS/MS and HPLC-ESI-MSn. The herbal drops (Crataegutt Tropfen, 94 mg/mL, extracting agent: Ethanol 45%) was diluted 10 times in methanol and directly analyzed.
UPLC- Q-TOF-MS/MS
The UPLC system (Agilent infinity 1260 series, Germany) was incorporated a binary pump, an auto sampler (G1367E), degasser (G1322A) and a DAD detector (1315D) with a light-pipe flow cell (recording at 280 and 320 nm). This was coupled to Ultra-Highresolution- Quadropole-Time-of-Flight (UHR-Q-TOF) (Bruker Impact HD, Bruker DaltoniK GmbH, Bremen Germany) equipped with an ESI source operating on Auto-MS/MS mode. The analysis was achieved in the negative ion mode in a mass range from m/z 50-1200. The ESI source parameters were: capillary voltage 4.5 KV; nebulising gas pressure 1.8 Bar; drying gas temperature 200.0°C, drying gas flow 9.0 L/min; Funnel 1RF 250.0 Vpp; transfer time 50.0 μs; and pre-pulse storage 2.0 μs. The MS data were analyzed through Data Analysis 4.2 software (Bruker Daltonics, Bremen, Germany). Internal calibration was achieved with 10 mL of 0.1 M sodium formate solution injected through a six port valve prior to each chromatographic run. Calibration was done using the High Precision Calibration (HPC).
UPLC and HPLC
The UPLC separation was achieved on a Polaris reverse phase C18 amide (RF-C18-A), 150 length x 2 mm inner-diameter, particle size 3 μm column (Agilent, Germany). Solvent A was water : formic acid (1000 : 0.05 v/v) and solvent B was methanol. Solvents were delivered at a total flow rate of 0.2 mL/min. The gradient profile was from 10% B to 80% B linearly in 70 min followed by 10 min isocratic and a return to 10% B at 90 min and 10min isocratic to re-equilibrate. The injection volume was 2 μL.
The HPLC separation was achieved on a 250 length x 3 mminner- diameter column containing 5 μm C18 amide, with a 5 mm x 3 mm-inner-diameter guard column (Varian, Darmstadt, Germany). Solvent A was water : formic acid (1000 : 0.05 v/v), and solvent B was acetonitrile. Solvents were delivered at a total flow rate of 0.5 mL/ min. The gradient profile was from 6% B to 80% B linearly in 70 min followed by 10 min isocratic and a return to 10% B at 90 min and 10 min isocratic to re-equilibrate. The injection volume was 5 μL [22,23].
HPLC-MSn
The HPLC-MSn analysis was achieved as previously described [21- 23].
UV Irradiation
UV irradiation experiments were performed as previously reported [24].
Results and Discussion
A reversed phase UHPLC-MS method was developed, allowing separation of 113 peaks in methanolic Crataegus extracts. Compound assignment was carried out for the identified phytoconstituents along with their m/z experimental and calculated, error (ppm), MS/MS fragments and molecular formula is presented in Table 1. Chemical structures of the identified phytoconstituents are shown in Figure 1. All identified compounds displaying a mass error of below 2 ppm thus confirming their elemental composition. Only three compounds errors were higher than 2 and below 5 ppm but their MS/MS and tandem MS data were with agreement with molecular formulas and suggested structures. Moreover, in this study not all identified compounds were fragmented in the Q-TOF. Consequently, the tandem MS fragmentation obtained from the LC-MSn was considered using a quadruple ion trap MS detector (Table 2). The UV chromatograms at 280 nm are shown in Figure S1 in the Supporting Information. Furthermore, compounds were also compared with authentic reference standards, structured under the same experimental conditions. Taking into account the data previously reported in literature, the flavonol glycosides were allowed further structure assignment and differentiated by using suggested rules and identification criteria previously reported [25].
| NO. | COmpOund Identity | MOl. FOrmula | The. m/z [M−H] | Exp. m/z [M−H] | Err. [ppm] | MS/MS fragments |
|---|---|---|---|---|---|---|
| 1 | Quinic acid | C7H12O6 | 191.0561 | 191.0561 | 0.1 | 173.0453, 127.0400, 111.0451, 85.0297 |
| 2 | 3-O-CaffeOylquinic acid | C16H18O9 | 353.0878 | 353.0875 | 1.0 | 191.0562, 179.0350, 173.0455, 133.0296 |
| 3 | cis-3-O-CaffeOylquinic acid | C16H18O9 | 353.0878 | 353.0875 | 0.9 | 191.0561, 179.0350, 177.0811, 173.0446, 135.0447 |
| 4 | 5-O-CaffeOylquinic acid | C16H18O9 | 353.0878 | 353.0872 | 1.8 | 191.0559, 179.0342, 173.0449 |
| 5 | cis- 5-O-Caffeoylquinic acid | C16H18O9 | 353.0878 | 353.0873 | 1.6 | 191.0549, 179.0347, 133.0301 |
| 6 | 3-O-(4'-O-Caffeoyl glucosyl)quinic acid | C22H28O14 | 515.1406 | 515.1403 | 0.6 | Not fragmented |
| 7 | 4-O-(4'-O-Caffeoyl glucosyl)quinic acid | C22H28O14 | 515.1406 | 515.1406 | 0.5 | Not fragmented |
| 8 | 5-O-(3'-O-Caffeoyl glucosyl)quinic acid | C22H28O14 | 515.1406 | 515.1403 | 0.6 | 179.0350, 191.0541 |
| 9 | 5-O-(4'-O-Caffeoyl glucosyl)quinic acid | C22H28O14 | 515.1406 | 515.1399 | 1.4 | 323.0811, 191.0563, 96.9699 |
| 10 | cis 5-O-(3'-O-Caffeoyl glucosyl)quinic acid | C22H28O14 | 515.1406 | 515.1407 | 0.0 | Not fragmented |
| 11 | Di-O-glycosyl-glucitol | C18H34O16 | 505.1774 | 505.1772 | 0.5 | Not fragmented |
| 12 | 3-O-p-Coumaroylquinic acid | C16H18O8 | 337.0929 | 337.0928 | 0.3 | 163.0401, 119.0501 |
| 13 | cis-3-O-p-Coumaroylquinic acid | C16H18O8 | 337.0929 | 337.0927 | 0.6 | 191.0558, 173.0449, 163.0398, 93.0346 |
| 14 | 4-O-p-Coumaroylquinic acid | C16H18O8 | 337.0929 | 337.0925 | 1.1 | 173.0455, 163.0395, 93.0364 |
| 15 | 5-O-p-Coumaroylquinic acid | C16H18O8 | 337.0929 | 337.0925 | 1.0 | 191.0542, 163.0397, 119.0501 |
| 16 | cis-5-O-p-Coumaroylquinic acid | C16H18O8 | 337.0929 | 337.0923 | 1.8 | 191.0551, 173.0452, 163.0398, 119.0494 |
| 17 | (Epi)catechin-4,8′-(epi)catechinC-hexoside | C36H36O17 | 739.1880 | 739.1885 | -0.7 | Not fragmented |
| 18 | (Epi)catechin-4,8′-(epi)catechinC-hexoside | C36H36O17 | 739.1880 | 739.1880 | 0.4 | Not fragmented |
| 19 | p-Coumaric acidO-hexoside | C15H18O8 | 325.0929 | 325.0927 | 0.7 | 163.0401, 119.0503 |
| 20 | (Epi)catechinC-hexoside | C21H24O11 | 451.1246 | 451.1241 | 1.0 | 331.0820, 290.0922, 272.0797 |
| 21 | CinchonainIa (isomer) | C24H20O9 | 451.1035 | 451.1030 | 1.0 | 341.0658, 315.0866, 289.0709, 287.0553, 217.0133 |
| 22 | CinchOnainIa (isOmer) | C24H20O9 | 451.1035 | 451.1035 | -0.1 | 341.0659, 287.0562, 217.0142, 161.0245, 112.9856 |
| 23 | Benzyl alcOhOl-hexOse-pentOse | C18H26O10 | 401.1453 | 401.1455 | -0.3 | 269.1041 |
| 24 | Quercetin 3,7-di-O-hexOside | C27H30O17 | 625.1410 | 625.1411 | -0.1 | NOt fragmented |
| 25 | (Epi)catechin-4,8′-(epi)catechin (isOmer) | C30H26O12 | 577.1351 | 577.1348 | 0.6 | 425.0883, 289.0704, 331.0803, 245.0461, 125.0239 |
| 26 | (Epi)catechin-4,8′-(epi)catechin (isOmer) | C30H26O12 | 577.1351 | 577.1347 | 0.7 | 425.0865, 407.0762, 451.1021, 289.0711, 245,0450 |
| 27 | (Epi)catechin-4,8′-(epi)catechin (isOmer) | C30H26O12 | 577.1351 | 577.1350 | 0.3 | 289.0729, 287.0585, 161.0260, 125.0249 |
| 28 | LuteOlin 6,8-di-C-hexOside (isOmer) | C27H30O16 | 609.1461 | 609.1459 | 0.3 | NOt fragmented |
| 29 | LuteOlin 6,8-di-C-hexOside (isOmer) | C27H30O16 | 609.1461 | 609.1462 | -0.2 | NOt fragmented |
| 30 | Epicatechin | C15H14O6 | 289.0718 | 289.0716 | 0.6 | 245.0825, 200.0570, 128.0356 |
| 31 | Cyanidin 7-O-glucOside | C21H21O11+ | 449.1089 | 449.1082 | 1.6 | 287.0554, 259.0613, 125.0230 |
| 32 | Cyanidin 3-O-galctOside | C21H21O11+ | 449.1089 | 449.1085 | 0.9 | 287.0558, 151.0037 |
| 33 | Cyanidin 3-O-glucoside | C21H21O11+ | 449.1089 | 449.1082 | 1.7 | 287.0575, 341.0648, 112.9842 |
| 34 | Apigenin 6,8-di-C-galactoside (vicenin-4) | C27H30O15 | 593.1512 | 593.1507 | 0.8 | 133.0143, 96.9706 |
| 35 | Apigenin 6,8-di-C-glucoside (vicenin-3) | C27H30O15 | 593.1512 | 593.1503 | 1.5 | 293.0435, 159.0 297 |
| 36 | 5-O-Feruloylquinic acid | C17H20o9 | 367.1035 | 367.1034 | 0.1 | 191.0559, 173.0450, 111.0474, 93.0339 |
| 37 | Apigenin 6-C-pentOsyl-8-C-hexOside (isomer) | C26H28O14 | 563.1406 | 563.1405 | 0.3 | 133.0143, 96.9695 |
| 38 | Apigenin 6-C-pentosyl-8-C-hexoside (isomer) | C26H28O14 | 563.1406 | 563.1403 | 0.5 | 357.1313, 164.9578, 133.0138, 96.9649 |
| 39 | Apigenin 6-C-hexosyl-8-C-pentoside (isomer) | C26H28O14 | 563.1406 | 563.1403 | 0.5 | 443.0980, 413.0861, 293.0446 |
| 40 | Apigenin 6-C-pentosyl-8-C-hexoside (isomer) | C26H28O14 | 563.1406 | 563.1407 | -0.1 | 443.0974, 413.0872, 293.0447, 133.0143 |
| 41 | 3-O-Caffeoylshikimic acid | C16H16O8 | 335.0772 | 335.0773 | -0.1 | 133.0301, 93.0318 |
| 42 | (Epi)catechin-4,8′-(epi)catechin-4′,8″-(epi)catechin (isomer) | C45H38O18 | 865.1985 | 865.1947 | 4.5 | Not fragmented |
| 43 | (Epi)catechin-4,8′-(epi)catechin-4′,8″-(epi)catechin (isomer) | C45H38O18 | 865.1985 | 865.1976 | 1.1 | Not fragmented |
| 44 | p-Coumaric acid | C9H8O3 | 163.0401 | 163.0401 | -0.4 | 119.0502, 96.9615 |
| 45 | Eriodictyol-di-C-hexoside | C27H32O16 | 611.1618 | 611.1615 | 0.3 | Not fragmented |
| 46 | (Epi)afzelechin-(epi)catechin | C30H26O11 | 561.1402 | 561.1402 | 0.1 | 289.0715, 271.0604, 125.0237, 96.9695 |
| 47 | Luteolin 8-C-galactoside | C21H20O11 | 447.0933 | 447.0929 | 0.9 | 327.0505, 357.0611, 297.0392, 163.0760 |
| 48 | Luteolin 8-C-glucoside (orientin) | C21H20O11 | 447.0933 | 447.0932 | 0.1 | 357.0609, 327.0506, 297.0398, 285.0368 |
| 49 | NaringeninC-hexoside (isomer) | C21H22O10 | 433.1140 | 433.1136 | 1.0 | 313.0708, 343.0835, 161.0458, 96.9694 |
| 50 | NaringeninC-hexoside (isomer) | C21H22O10 | 433.1140 | 433.1138 | 0.6 | 343.0830, 313.0720, 96.9688, 78.9593 |
| 51 | Quercetin 3-O-(2, 6-di-O-rhamnosyl-glucoside) | C33H40O20 | 755.2040 | 755.2037 | 0.4 | Not Fragmented |
| 52 | Apigenin 8-C-glucoside (vitexin) | C21H20O10 | 431.0984 | 431.0982 | 0.3 | 311.0557, 341.0659, 284.0621 |
| 53 | Apigenin 6-C-glucoside (isovitexin) | C21H20O10 | 431.0984 | 431.0985 | 0.3 | 311.0560, 341.0663, 413.0868, 191.0560, 283.0607, 296.0456 |
| 54 | Isorhamnetin 7-O-(6-O-rhamnosyl-glucoside) | C28H32O16 | 623.1618 | 623.1619 | -0.2 | 150.0310, 112.9861, 96.9681, 78.9583, 293. 0461 |
| 55 | Isorhamnetin 3-O-(6-O-rhamnosyl-galactoside) | C28H32O16 | 623.1618 | 623.1601 | 2.7 | 112.9858 |
| 56 | Isorhamnetin 3-O-(6-O-rhamnosyl-glucoside) | C28H32O16 | 623.1618 | 623.1616 | 0.2 | Not fragmented |
| 57 | Kaempferol 7-O-(6-O-rhamnosyl-glucoside) | C27H30O15 | 593.1512 | 593.1517 | -0.9 | 159.0296, 112.9848 |
| 58 | Kaempferol 3-O-(6-O-rhamnosyl-galactoside) | C27H30O15 | 593.1512 | 593.1511 | 0.1 | 151.0411, 112.9856 |
| 59 | Kaempferol 3-O-(6-O-rhamnosyl-glucoside) | C27H30O15 | 593.1512 | 593.1512 | 0.2 | Not fragmented |
| 60 | Diosmetin 7-O-rutinoside (diosmin) | C28H32O15 | 607.1668 | 607.1665 | 0.5 | Not fragmented |
| 61 | Kaempferol 3-O-glucuronide | C21H18O12 | 461.0725 | 461.0725 | 0.2 | Not fragmented |
| 62 | Luteolin 7-O-glucoside | C21H20O11 | 447.0933 | 447.933 | 0.0 | 285.0406, 249.0532, 174.09551 |
| 63 | Kaempferol 7-O-glucoside | C21H20O11 | 447.0933 | 447.0930 | 0.7 | 284.0323, 174.9585, 242.9443 |
| 64 | Kaempferol 3-O-glucoside | C21H20O11 | 477.0933 | 447.0927 | 1.4 | 285.0393, 256.0357, 174.9558 |
| 65 | Quercetin 3-O-(6-O-rhamnosyl-glucoside) (rutin) | C27H30O16 | 609.1461 | 609.1458 | 0.6 | 301.0351, 187.0974, 111.0060 |
| 66 | Myricetin 3-O-rhamnoside (myricitrin) | C21H20O12 | 463.0882 | 463.0880 | 0.4 | 316.0216, 178.9982, 151.0037 |
| 67 | Quercetin 3-O-galactoside | C21H20O12 | 463.0982 | 463.0878 | 0.8 | 301.0342, 300.0270, 273.0054, 178.9968, 151.0038 |
| 68 | Quercetin 3-O-glucoside | C21H20O12 | 463.0882 | 463.0876 | 1.2 | 301.0347 |
| 69 | 3,4-di-O-Caffeoylquinic acid | C25H24O12 | 515.1195 | 515.1193 | 0.4 | Not fragmented |
| 70 | 3,5-di-O-Caffeoylquinic acid | C25H24O12 | 515.1195 | 515.1194 | 0.3 | Not fragmented |
| 71 | 4,5-di-O-Caffeoylquinic acid | C25H24O12 | 515.1195 | 515.1192 | 0.5 | 353.0898, 179.0377, 143.1068, 126.9994 |
| 72 | IsorhamnetinO-hexoside | C22H22O12 | 477.1038 | 477.1036 | 0.6 | 315.0502, 300.0264 |
| 73 | Naringenin 7-O-glucoside | C21H22O10 | 433.1140 | 433.1139 | 0.3 | 271.0611, 151.0032, 227.0715 |
| 74 | Apigenin 7-O- rutinoside | C27H30o14 | 577.1563 | 577.1557 | 1.0 | 296.0466, 112.9857, 96.9693 |
| 75 | Quercetin 3-O-xyloside | C20H18O11 | 433.0776 | 433.0772 | 1.0 | 300.0273, 301.0334, 167.0326, 149.0235 |
| 76 | Apigenin 7-O-glucoside | C21H20O10 | 431.0984 | 431.0979 | 1.0 | 269.0435, 173.0443, 149.0222, 96.9676 |
| 77 | Quercetin 3-O-rhamnoside (quercitrin) | C21H20O11 | 447.0933 | 447.0928 | 0.4 | 300.0268, 301.0330, 271.0244, 255.0290, 151.0031 |
| 78 | Pectolinarin | C29H34O15 | 621.1825 | 621.1827 | -0.2 | 313.0754, 295.0595, 175.0040 |
| 79 | QuercetinO-acetyl hexoside (isomer) | C23H22O13 | 505.0988 | 505.0984 | 0.3 | 300.0248, 301.0283, 271.0262, 255.0239, 151.0027 |
| 80 | QuercetinO-acetyl hexoside (isomer) | C23H22O13 | 505.0988 | 505.0982 | 0.5 | 300.0274, 271.0260, 255.0307, 151.0014 |
| 81 | Phloretin 2'-O-glucoside (phlorizin) | C21H24O10 | 435.1297 | 435.1294 | 0.7 | 273.0781 |
| 82 | (−)-11-hydroxy-9,10-dihydrojasmonic acid 11-β-D-glucoside | C18H30O9 | 389.1817 | 389.1812 | 0.7 | 183.1376, 134.0263, 101.0216, 227.1339 |
| 83 | Kaempferol 3-O-rhamnoside | C21H20O10 | 431.0984 | 431.0980 | 0.9 | Not fragmented |
| 84 | Cyanidin | C15H11O6+ | 287.0561 | 287.0561 | 0.0 | 151.0017, 135.0446, 107.0153 |
| 85 | 3-O-Methylellagic acid 4'-(2'',3''-di-O-acetyl)-rhamnoside | C25H22O14 | 545.0937 | 545.0920 | 1.7 | 299.0190, 314.0443 |
| 86 | Quercetin | C15H10O7 | 301.0354 | 301.0349 | 0.5 | 151.0021, 130.9922 |
| 87 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin(isomer) | C45H36O18 | 863.1829 | 863.1843 | -1.6 | Not fragmented |
| 88 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin(isomer) | C45H36O18 | 863.1829 | 863.1839 | -1.2 | Not fragmented |
| 89 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin (isomer) | C45H36O18 | 863.1829 | 863.1855 | -3.1 | Not fragmented |
| 90 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin(isomer) | C45H36O18 | 863.1829 | 863.1847 | -2.1 | NOt fragmented |
| 91 | PrOtOcatechuic acid O-hexOside | C13H16O9 | 315.0772 | 315.0770 | 0.5 | 153.0184, 152.0113, 109.0288, 108.0213 |
| 92 | Citric acid | C6H8O7 | 191.0197 | 191.0198 | -0.2 | 102.9491, 85.0296 |
| 93 | Syringic acid O- hexOside | C15H20O10 | 359.0984 | 359.0988 | -0.6 | 197.0440, 175.0063, 153.0556, 149.0230 |
| 94 | Salicylic acid O-galactOside | C13H16O8 | 299.0772 | 299.0774 | -0.5 | 137.0245, 93.0347, 239.0560, 179.0353 |
| 95 | Salicylic acid O-glucOside | C13H16O8 | 299.0772 | 299.0773 | -0.1 | 137.0237, 93.0346 |
| 96 | HOmOvanillic acid O-hexOside | C15H20O9 | 343.1035 | 343.1034 | 0.1 | 181.0510, 163.0396, 119.0494 |
| 97 | Sinapic acid O-hexOside | C17H22O10 | 385.1140 | 385.1137 | 0.9 | 223.0618, 179.0698 |
| 98 | Salicylic acid | C7H6O3 | 137.0244 | 137.0244 | -0.2 | 93.0347 |
| 99 | PhlOretin | C15H14O5 | 273.0768 | 273.0767 | 0.1 | NOt fragmented |
| 100 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin (isOmer) | C30H24O12 | 575.1195 | 575.1196 | -0.2 | 423.0742, 289.0718, 285.0403, 125.0242 |
| 101 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin (isOmer) | C30H24O12 | 575.1195 | 575.1191 | 0.8 | 449.0821, 423.0782, 289.0720, 285.0400, 245.0462, 152.0249 |
| 102 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin (isOmer) | C30H24O12 | 575.1195 | 575.1192 | 0.6 | 449.0864, 407.0818, 327.0480, 289.0706, 285.0401, 125.0245 |
| 103 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin (isOmer) | C30H24O12 | 575.1195 | 575.1192 | 0.5 | 287.0546, 125.0242 |
| 104 | Vanillic acid O-hexOside | C14H18O9 | 329.0878 | 329.0873 | 0.9 | 167.0345, 152.0140, 123.0451, 108.0230 |
| 105 | Syringicacid | C9H10O5 | 197.0455 | 197.0457 | -0.7 | NOt fragmented |
| 106 | Malic acid | C4H6O5 | 133.0142 | 133.0142 | 0.2 | 115.0034 |
| 107 | Pyruvic acid | C3H4O3 | 87.0088 | 87.00 | 0.2 | NOt fragmented |
| 108 | AscOrbic acid | C6H8O6 | 175.0248 | 175.0248 | -0.0 | 87.0090 |
| 109 | Cratenacin (isOmer) | C29H32O15 | 619.1668 | 619.1668 | 0.1 | 413.0874, 293.0458 |
| 110 | Cratenacin (isOmer) | C29H32O15 | 619.1668 | 619.1667 | 0.2 | 413.0872, 293.0447, 499.1215, 353.0709, 577.1545 |
| 111 | Cratenacin (isOmer) | C29H32O15 | 619.1668 | 619.1668 | -0.0 | 413.0871, 293.0452, 499.1244 |
| 112 | Vitexin 2″-O-rhamnOside | C27H30O14 | 577.1563 | 577.1561 | 0.3 | 413.0872, 457. 1133, 293, 0457 |
| 113 | IsOvitexin 2″-O-rhamnOside | C27H30O14 | 577.1563 | 577.0561 | 0.2 | 413.0870, 293.0456, 457.1128 |
Table 1: UPLC-QTOf-MS/MS mass spectral data in negative iOn mOde Of C. laevigata, C. mOnOgyna and the herbal drOps (Crataegutt TrOpfen) phenOlics.
| No. | Compound Identity | tR(min) | [M−H] | Characteristic m/z of ions in negative ion mode | Herbal drug | C. laevigata | C. monogyna | ||
|---|---|---|---|---|---|---|---|---|---|
| F | L | F | L | ||||||
| 1 | Quinic acid | 2.9 | 191 | MS22→ 173 (100), 127 (97), 111 (37), 85 (68); MS23→ 127 (96), 111 (100) | P | P | P | P | P |
| 2 | 3-O-Caffeoylquinic acid | 12.6 | 353 | MS22→ 191 (100), 179 (49); MS23→ 127 (100), 173 (81), 85 (78) | P | P | P | P | - |
| 3 | cis-3-O-Caffeoylquinic acid | 13.4 | 353 | MS22→ 191 (100), 179 (42); MS23→ 127 (100), 173 (35), 85 (31) | P | P | - | - | - |
| 4 | 5-O-Caffeoylquinic acid | 20.0 | 353 | MS22→ 191 (100); MS23→ 173 (100), 127 (75), 111 (53), 85 (98); MS24→ 93 (100), 109 (37) | P | P | P | P | P |
| 5 | cis- 5-O-Caffeoylquinic acid | 23.2 | 353 | MS22→ 191 (100); MS23→ 127 (100), 173 (32), 85 (63) | P | P | - | P | - |
| 6 | 3-O-(4'-O-Caffeoyl glucosyl)quinic acid | 11.8 | 515 | MS22→ 353 (100), 191 (30), 341 (28), 323 (04), 335 (07), 179 (11); MS23→ 191 (100), 179 (09); MS24→ 127 (100), 173 (85), 157 (39), 85 (39), 111 (72) | P | - | P | P | P |
| 7 | 4-O-(4'-O-Caffeoyl glucosyl)quinic acid | 13.5 | 515 | MS22→ 341 (100), 353 (94), 173 (67), 179 (83), 323 (01); MS23→ 173 (100), 179 (52), 191 (22) | P | - | - | - | - |
| 8 | 5-O-(3'-O-Caffeoyl glucosyl)quinic acid | 16.5 | 515 | MS22→ 323 (100), 353 (30), 341 (13), 191 (34), 161 (10); MS23→ 161 (100), 133 (10) | P | - | P | P | P |
| 9 | 5-O-(4'-O-Caffeoyl glucosyl)quinic acid | 18.1 | 515 | MS22 → 353 (100), 341 (94), 395 (25), 323 (15), 191 (79), 179 (36); MS23→ 191 (100), 179 (09) | - | - | - | - | P |
| 10 | cis 5-O-(3'-O-Caffeoyl glucosyl)quinic acid | 22.2 | 515 | MS22→ 323 (100), 353 (38), 341 (10), 191 (155); MS23→ 161 (100), 133 (04), 323 (99); MS24→ 161 (100), 117(99) | - | - | - | - | P |
| 11 | Di-O-glycosyl-glucitol | 12.1 | 505 | MS22→ 343 (100), 181 (25); MS23→ 181 (100) | P | - | P | - | - |
| 12 | 3-O-p-Coumaroylquinic acid | 17.1 | 337 | MS22→ 163 (100), 191 (12); MS23→ 119 (100) | P | P | - | P | - |
| 13 | cis-3-O-p-Coumaroylquinic acid | 17.8 | 337 | MS22→ 163 (100), 191 (07); MS23→ 119 (100) | P | P | - | P | P |
| 14 | 4-O-p-Coumaroylquinic acid | 26.1 | 337 | MS22→ 173 (100), 163 (10); MS23→ 93 (100), 155 (29), 111 (65), 71 (51) | P | P | - | P | - |
| 15 | 5-O-p-Coumaroylquinic acid | 27.8 | 337 | MS22→ 191 (100), 163 (10);MS23→ 127 (100), 173 (58), 85 (74), 109 (23); MS24→ 109 (100), 81 (15) | P | P | P | - | - |
| 16 | cis-5-O-p-Coumaroylquinic acid | 29.7 | 337 | MS22→ 191 (100), 163 (06);MS23→ 127 (100), 173 (38), 85 (29), 109 (27), 93 (13) | P | p | P | P | P |
| 17 | (Epi)catechin-4,8′-(epi)catechinC-hexoside | 17.3 | 739 | MS22→ 449 (100), 587 (57), 331 (56), 288 (19);MS23→ 327 (100), 289 (71), 245 (28), 167 (18) | P | - | - | - | - |
| 18 | (Epi)catechin-4,8′-(epi)catechinC-hexoside | 21.8 | 739 | MS22→ 449 (100), 289 (15), 467 (46), 649 (62), 619 (68), 587 (40), 329 (52), 299 (12); MS23→ 329 (100), 289 (26), 245 (10), 287 (26); MS24→ 167 (100), 299 (49) | P | - | - | - | - |
| 19 | p-Coumaric acidO-hexoside | 17.6 | 325 | MS22→ 163 (100), 119 (15); MS23→ 119 (100) | P | - | P | P | P |
| 20 | (Epi)catechinC-hexoside | 18.3 | 451 | MS22→ 331 (100), 361 (21), 289 (12); MS23→ 313 (100), 287 (41), 269 (29), 245 (20); MS24→ 231 (100), 245 (15), 203 (46) | P | P | - | - | - |
| 21 | CinchonainIa (isomer) | 65.2 | 451 | MS22→ 313 (100), 289 (52), 331 (79), 169 (10), 137 (11); MS23→ 134 (100), 168 941), 269 (31), 295 (14); MS24→ 123 (100), 150 (89) | P | - | P | P | - |
| 22 | CinchonainIa (isomer) | 66.7 | 451 | MS22→ 289 (100), 331 (18); MS23→ 215 (100), 245 (79), 267 (43), 93 (17); MS24→ 106 (100), 146 (43), 172 (83), 197 (19) | P | - | P | P | - |
| 23 | Benzyl alcohol-hexose-pentose | 21.6 | 401 | MS22→ 269 (100) | - | P | P | P | P |
| 24 | Quercetin 3,7-di-O-hexoside | 21.9 | 625 | MS22→ 463 (100), 301 (42); MS23→ 301 (100), 300 (27), 271 (09); MS24→ 271 (98), 299 (100), 179 (18), 151 (19) | P | - | - | - | - |
| 25 | (Epi)catechin-4,8′-(epi)catechin (isomer) | 22.3 | 577 | MS22→ 407 (100), 425 (79), 451 (20), 289 (18),;MS23→ 285 (100), 389 (24), 297 (30), 255 (30), 255 (32), 243 (15) | P | P | - | - | - |
| 26 | (Epi)catechin-4,8′-(epi)catechin (isomer) | 25.4 | 577 | MS22→ 407 (100), 425 (68), 451 (22), 289 (22);MS23→ 285 (100), 281 (37), 389 (36), 255 (23), 423 (18); MS24→ 257 (100), 283 (84) | P | P | P | - | P |
| 27 | (Epi)catechin-4,8′-(epi)catechin (isomer) | 41.7 | 577 | MS22→ 407 (100), 425 (93), 451 (24), 287 (26); MS23→ 285 (100), 281 (60); MS24→ 255 (100) | - | P | P | - | - |
| 28 | Luteolin 6,8-di-C-hexoside (isomer) | 23.9 | 609 | MS22→ 489 (100), 519 (21), 591 (8), 429 (5), 399 (15), 369 (26);MS23→ 369 (100), 399 (52); MS24→ 341 (100), 351 (20), 313 (28), 298 (10) | P | - | - | - | - |
| 29 | Luteolin 6,8-di-C-hexoside (isomer) | 30.9 | 609 | MS22→ 429 (100), 489 (57), 339 (31), 357 (29), 327 (15), 309 (35);MS23→ 309 (100), 339 (26), 285 (03), 351 (55); MS24→ 309 (100), 267 (61), 238 (10), 176 (23), 172 (17) | P | - | - | - | - |
| 30 | Epicatechin | 25.8 | 289 | MS22→ 245 (100), 205 (33), 179 (11);MS23→ 203 (100), 227 (25), 187 (23), 161 (20) | P | P | P | P | - |
| 31 | Cyanidin 7-O-glucoside | 27.0 | 449 | MS22→ 287 (100), 269 (51), 259 (42); MS23→ 259 (100), 243 (16); MS24→ 725 (100), 215 (76), 173 (51), 241 (20) | P | P | - | P | - |
| 32 | Cyanidin 3-O-galctoside | 38.5 | 449 | MS22→ 287 (100), 269 (12), 151 (09);MS23→ 151 (100); MS24→ 107 (100) | P | P | P | P | - |
| 33 | Cyanidin 3-O-glucoside | 56.8 | 449 | MS22→ 287 (100), 407 (05), 341 (53), 243 (58);MS23 → 243 (100), 228 (16); MS24→ 228 (100), 122 (82) | P | - | - | - | P |
| 34 | Apigenin 6,8-di-C-galactoside (vicenin-4) | 27.1 | 593 | MS22→ 473 (100), 503 (24), 383 (35), 353 (69); MS23→ 353 (100), 383 (14); MS24→ 325 (100), 297 (56) | P | - | P | - | P |
| 35 | Apigenin 6,8-di-C-glucoside (vicenin-3) | 36.0 | 593 | MS22→ 413 (100), 473 (05), 293 (35);MS23→ 293 (100); MS24→ 293 (100), 249 (19), 174 (21) | P | - | P | - | P |
| 36 | 5-O-Feruloylquinic acid | 28.6 | 367 | MS22→ 191 (100); MS23→ 127 (100), 173 (32), 85 (55), 109 (20) | P | P | P | P | P |
| 37 | Apigenin 6-C-pentosyl-8-C-hexoside (isomer) | 28.7 | 563 | MS22→ 353 (100), 543 (34), 503 (33), 473 (66), 443 (43), 383 (88), 289 (26);MS23→ 325 (100), 297 (59); MS24→ 297 (100) | P | P | - | - | - |
| 38 | Apigenin 6-C-pentosyl-8-C-hexoside (isomer) | 30.6 | 563 | MS22→ 353 (100), 503 (43), 473 (99), 443 (78), 383 (72); MS23→ 325 (100), 353 (17), 297 (64); MS24→ 297 (100) | P | P | - | - | - |
| 39 | Apigenin 6-C-hexosyl-8-C-pentoside (isomer) | 32.2 | 563 | MS22→ 443 (100), 383 (52), 353 (66), 473 (54);MS23 → 325 (100), 353 (10), 297 (57); MS24→ 297 (100), 325 (09) | P | P | P | P | |
| 40 | Apigenin 6-C-pentosyl-8-C-hexoside (isomer) | 35.0 | 563 | MS22→ 353 (100), 473 (68), 443 (38), 413 (43), 383, (56), 503 (15);MS23→ 293 (100); MS24→ 293 (100), 249 (36), 221 (11), 173 (26) | P | - | P | - | - |
| 41 | 3-O-Caffeoylshikimic acid | 29.7 | 335 | MS22→ 191 (100), 179 (70), 135 (19), 173 (9);MS23→ 133 (100) | P | P | - | - | - |
| 42 | (Epi)catechin-4,8′-(epi)catechin-4′,8″-(epi)catechin (isomer) | 26.5 | 865 | MS22 → 695 (100), 577 (70), 739 (30), 713 (39), 449 (31), 425 (30), 407 (05), 287 (47); MS23→ 525 (100), 677 (49), 543 (31), 451 (64), 407 (83), 243 (53) | - | - | P | - | P |
| 43 | (Epi)catechin-4,8′-(epi)catechin-4′,8″-(epi)catechin (isomer) | 31.2 | 865 | MS22→ 695 (100), 577 (56), 575 (23), 739 (32), 711 (32), 559 (14), 543 (31), 451 (30), 425 (46), 407 (64), 289 (22), 287 (31);MS23→ 525 (100), 543 (95), 677 (53), 651 (12), 451 (33), 407 (44), 363 (25), 289 (13), 243 (53); MS24→ 525 (100), 391 (10) | P | P | P | - | P |
| 44 | p-Coumaric acid | 32.1 | 163 | MS22→ 119 (100) | P | - | - | - | - |
| 45 | Eriodictyol-di-C-hexoside | 32.4 | 611 | MS22→ 431 (100), 449 (04), 327 (06), 251 (38);MS23→ 251 (100), 309 (39), 207 (16); MS24→ 207 (100), 189 (56) | P | P | P | P | P |
| 46 | (Epi)afzelechin-(epi)catechin | 32.7 | 561 | MS22→ 289 (100), 407 (20), 425 (19), 435 (41), 543 (22), 329 (17), 271 (16);MS23→ 245 (100), 205 (36), 179 (17); MS24→ 203 (100), 226 (20), 188 (12) | P | P | - | - | - |
| 47 | Luteolin 8-C-galactoside | 33.4 | 447 | MS22→ 327 (100), 357 (77);MS23→ 299 (100), 327 (21), 284 (11); MS24→ 299 (100), 282 (33), 271 (26), 255 (100), 213 (40) | P | P | P | P | - |
| 48 | Luteolin 8-C-glucoside (orientin) | 34.5 | 447 | MS22→ 327 (100), 357 (81), 429 (17);MS23→ 299 (100), 284 (16), 285 (05); MS24→ 213 (100), 271 (15), 175 (28) | P | P | - | - | - |
| 49 | NaringeninC-hexoside (isomer) | 34.1 | 433 | MS22→ 313 (100), 343 (18); MS23→ 193 (100), 206 (10), 167 (17); MS24→ 165 (100), 149 (57), 125 (44), | P | P | P | - | P |
| 50 | NaringeninC-hexoside (isomer) | 35.2 | 433 | MS22→ 313 (100), 343 (28); MS23→ 193 (100), 207 (11), 167 (15); MS24→ 165 (100), 149 (99), 137 (29), 125 (31) | P | - | P | - | P |
| 51 | Quercetin 3-O-(2, 6-di-O-rhamnosyl-glucoside) | 36.5 | 755 | MS22→ 300 (100), 301 (64), 609 (22), 591 (22), 489 (36), 343 (23), 737 (09), 271 (23), 255 (13); MS23→ 271 (100), 255 (56), 179 (16), 151 (15); MS24→ 271 (100), 255 (19), 243 (23), 267 (15), 151 (12) | P | - | - | - | - |
| 52 | Apigenin 8-C-glucoside (vitexin) | 37.9 | 431 | MS22→ 311 (100), 283 (10), 341 (06);MS23→ 283 (100); MS24→ 163 (100), 283 (20), 239 (37), 197 (17), 211 (12) | P | P | P | - | P |
| 53 | Apigenin 6-C-glucoside (isovitexin) | 41.4 | 431 | MS22→ 311 (100), 341 (45);MS23→ 283 (100); MS24 → 283 (100), 239 (46), 224 (36), 183 (28), 162 (34) | P | P | P | - | P |
| 54 | Isorhamnetin 7-O-(6-O-rhamnosyl-glucoside) | 40.2 | 623 | MS22→ 299 (100), 300 (30), 314 (79), 315 (47), 477 (11), 271 (26), 177 (25);MS23→ 271 (100), 255 (33), 243 (10) | P | - | - | - | - |
| 55 | Isorhamnetin 3-O-(6-O-rhamnosyl-galactoside) | 47.1 | 623 | MS22→ 315 (100), 300 (28), 271 (13);MS23→ 300 (100); MS24→ 271 (100), 255 (79) | P | - | - | - | - |
| 56 | Isorhamnetin 3-O-(6-O-rhamnosyl-glucoside) | 48.6 | 623 | MS22→ 315 (100), 300 (25), 271 (14);MS23→ 300 (100); MS24→ 271 (100), 255 (56), 221 (51) | P | - | - | - | - |
| 57 | Kaempferol 7-O-(6-O-rhamnosyl-glucoside) | 40.3 | 593 | MS22→ 285 (100);MS23→ 175 (100), 285 (76), 241 (86), 217 (56), 199 (91) | P | - | - | - | - |
| 58 | Kaempferol 3-O-(6-O-rhamnosyl-galactoside) | 45.3 | 593 | MS22→ 285 (100), 284 (28), 257 (11);MS23→ 255 (100), 239 (11), 151 (16) | P | - | - | - | - |
| 59 | Kaempferol 3-O-(6-O-rhamnosyl-glucoside) | 48.3 | 593 | MS22→ 284 (100), 463 (05);MS23→ 257 (100), 267 (54), 239 (41), 150 (30); MS24→ 109 (100), 163 (80), 212 (36) | P | - | P | - | P |
| 60 | Diosmetin 7-O-rutinoside (diosmin) | 40.6 | 607 | MS22→ 299 (100), 443 (27);MS23→ 271 (100), 255 (79), 179 (11), 151 (16) | P | - | - | - | - |
| 61 | Kaempferol 3-O-glucuronide | 40.9 | 461 | MS22→ 285 (100);MS23→ 199 (100), 285 (52), 241 (45), 226 (35), 175 (35), 155 (11); MS24→ 196 (100), 154 (64) | P | - | - | - | - |
| 62 | Luteolin 7-O-glucoside | 41.5 | 447 | MS22→ 285 (100);MS23→ 241 (100), 285 (72), 217 (55), 199 (77), 175 (85), 151 (52) | P | - | - | - | - |
| 63 | Kaempferol 7-O-glucoside | 48.3 | 447 | MS22→ 284 (100), 285 (95), 255 (22);MS23→ 255 (100); MS24→ 255 (100), 227 (85), 211 (27) | P | P | P | P | - |
| 64 | Kaempferol 3-O-glucoside | 50.5 | 447 | MS22→ 284 (100), 285 (20), 255 (20);MS23→ 255 (100), 227 (17); MS24→ 255 (100), 227 (68), 211 (36) | P | - | P | P | P |
| 65 | Quercetin 3-O-(6-O-rhamnosyl-glucoside) (rutin) | 41.9 | 609 | MS22→ 301 (100), 300 (32), 271 (15); MS23→ 151 (100), 179 (76), 255 (61), 300 (45), 179 (62), 107 (10) | P | P | P | P | P |
| 66 | Myricetin 3-O-rhamnoside (myricitrin) | 42.2 | 463 | MS22→ 316 (100), 317 (49);MS23→ 271 (100), 287 (32), 179 (32), 151 (13); MS24→ (100) | P | - | - | - | - |
| 67 | Quercetin 3-O-galactoside | 44.0 | 463 | MS22→ 301 (100), 300 (25);MS23→ 179 (100), 151 (90), 271 (50), 255 (35); MS24→ 151 (100) | P | P | P | P | P |
| 68 | Quercetin 3-O-glucoside | 45.0 | 463 | MS22→ 301 (100); MS23 → 179 (100), 151 (87), 255 (25), 271 (48); MS23→ 151 (100), 107 (15) | P | P | P | P | P |
| 69 | 3,4-di-O-Caffeoylquinic acid | 45.6 | 515 | MS22→ 353 (100), 173 (23), 179 (15);MS23→ 173 (100), 178 (56), 191 (51), 135 (10); MS24→ 111 (100), 93 (74), 71 (21), 155 (53) | P | - | - | - | - |
| 70 | 3,5-di-O-Caffeoylquinic acid | 47.0 | 515 | MS22→ 353 (100), 191 (08);MS23→ 191 (100), 179 (18), 135 (10); MS24→ 93 (100), 173 (90), 127 (86), 111 (47), 85 (87) | P | P | - | - | - |
| 71 | 4,5-di-O-Caffeoylquinic acid | 51.0 | 515 | MS22→ 353 (100);MS23→ 173 (100), 191 (28), 179 (62); MS24→ 93 (100), 111 (67), 155 (22) | P | - | - | P | - |
| 72 | IsorhamnetinO-hexoside | 45.9 | 477 | MS22→ 315 (100), 300 (73);MS23→ 300 (100); MS24→ 271 (100), 255 (99) | P | - | - | - | - |
| 73 | Naringenin 7-O-glucoside | 46.5 | 433 | MS22→ 271 (100);MS23→ 151 (100), 177 (26); MS24→ 107 (100) | P | - | P | - | P |
| 74 | Apigenin 7-O- rutinoside | 47.0 | 577 | MS22→ 269 (100);MS23→ 269 (100), 227 (17), 200 (17),158 (18), 117 (12); MS24→ 223 (100), (90) | P | - | - | - | - |
| 75 | Quercetin 3-O-xyloside | 47.6 | 433 | MS22→ 301 (100), 300 (25);MS23→ 151 (100), 271 (41), 179 (93); MS24→ 151 (100) | P | - | - | - | - |
| 76 | Apigenin 7-O-glucoside | 49.1 | 431 | MS22→ 269 (100), 301 (87);MS23→ 268 (100), 269 (64), 225 (17), 197 (16); MS24→ 197 (100) | P | - | - | - | - |
| 77 | Quercetin 3-O-rhamnoside (quercitrin) | 51.0 | 447 | MS22→ 301 (100), 300 (23);MS23→ 178 (100), 151 (91), 271 (42), 255 (28); MS24→ 107 (100), 169 (84) | P | - | - | - | - |
| 78 | Pectolinarin | 51.8 | 621 | MS22→ 313 (100), 343 (14), 413 (21), 501 (99);MS23 → 193 (100), 269 (04), 285 (01), 167 (17); MS24→ 165 (100), 149 (69), 137 (35), 81 (22) | P | - | P | - | - |
| 79 | QuercetinO-acetyl hexoside (isomer) | 52.2 | 505 | MS22→ 301 (100), 300 (37), 271 (08);MS23→ 271 (100), 255 (54), 179 (93), 151 (90), 107 (14); MS24→ 151 (100) | P | P | - | - | - |
| 80 | QuercetinO-acetyl hexoside (isomer) | 55.7 | 505 | MS22→ 300 (100), 301 (68);MS23→ 271 (100), 300 (49), 255 (52), 179 (58), 151 (53); MS24→ 271 (100), 255 (41), 243 (26) | P | - | - | - | - |
| 81 | Phloretin 2'-O-glucoside (phlorizin) | 53.9 | 435 | MS22→ 273 (100), 229 (21);MS23→ 229 (100), 107 (14); MS24→ 107 (100), 121 (43) | P | - | - | - | P |
| 82 | (−)-11-hydroxy-9,10-dihydrojasmonic acid 11-β-D-glucoside | 58.5 | 389 | MS22→ 227 (100), 183 (12);MS23→ 183 (100), 165 (91); MS24→ 112 (76), 163 (26) | P | - | - | - | - |
| 83 | Kaempferol 3-O-rhamnoside | 58.8 | 431 | MS22→ 284 (100), 285 (43);MS23→ 255 (100), 227 (23), 229 (13) | P | - | - | - | - |
| 84 | Cyanidin | 62.6 | 287 | MS22→ 151 (100), 135 (10);MS23→ 107 (100), 169 (10), 65 (20); MS24→ 65 (100) | P | - | - | - | - |
| 85 | 3-O-Methylellagic acid 4'-(2'',3''-di-O-acetyl)-rhamnoside | 64.8 | 545 | MS22→ 315 (100), 300 (68), 301 (12), 271 (21), 255 (27);MS23→ 300 (100), 301 (10); MS24→ 255 (100), 271 (67) | P | - | - | - | - |
| 86 | Quercetin | 69.9 | 301 | MS22→ 179 (100), 151 (83);MS23→ 151 (100), 169 (14); MS24→ 107 (100), 168 (14) | P | - | - | - | - |
| 87 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin (isomer) | 31.4 | 863 | MS22→ 711 (100), 695 (70), 577 (36), 451 (61), 407 (53), 287 (20), 575 (13), 425 (30) | - | P | - | - | - |
| 88 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin (isomer) | 32.0 | 863 | MS22→ 711 (100), 725 (41), 599 (39), 425 (23), 449 (15), 411(33), 287 (14); MS23→ 559 (100), 693 (98), 541 (59), 407 (42), 381 (25); MS24→ 567 (100), 658 (35), 407 (13) | - | P | - | - | - |
| 89 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin (isomer) | 33.8 | 863 | MS22→ 573 (100), 711 (50), 694 (25), 451 (51), 411) (82); MS23→ 411 (100), 273 (11); MS24→ 227 (100), 283 (24), 349 (45), 125 (14) | - | P | - | - | - |
| 90 | (Epi)catechin-(4,8′)-(epi)catechin-(4′,8″/2′,7″)-(epi)catechin (isomer) | 38.2 | 863 | MS22→ 711 (100), 693 (50), 599 (72), 407 (05); MS23→ 559 (100), 693 (82), 425 (10); MS24→ 567 (100), 483 (32), 405 (33), 282 (25) | - | P | - | - | - |
| 91 | Protocatechuic acid O-hexoside | 9.0 | 315 | MS22→ 153 (100), 109 (13); MS23→ 109 (100) | - | P | P | - | P |
| 92 | Citric acid | 4.2 | 191 | MS22→ 111 (100), 173 (22) | P | - | P | P | P |
| 93 | Syringic acid O-hexoside | 11.8 | 359 | MS22→ 197 (100), 182 (13); MS23→ 182 (100), 153 (23); MS24→ 167 (100), 123 (11) | P | P | P | P | P |
| 94 | Salicylic acid O-galactoside | 7.7 | 299 | MS22→ 137 (100); MS23→ 93 (100) | - | P | - | P | P |
| 95 | Salicylic acid O-glucoside | 11.3 | 299 | MS22→ 137 (100), 239 (70), 209 (19), 179 (73); MS23→ 93 (100) | P | - | - | P | P |
| 96 | Homovanillic acid O-hexoside | 17.6 | 343 | MS22→ 181 (100), 137 (11); MS23→ 137 (100); MS24→ 95 (100) | P | - | P | - | P |
| 97 | Sinapic acid O-hexoside | 20.8 | 385 | MS22→ 223 (100); MS23→ 208 (100), 179 (45), 165 (32); MS24→ 164 (100), 149 (22) | P | - | - | P | P |
| 98 | Salicylic acid | 7.6 | 137 | MS22→ 93 (100) | P | - | - | - | - |
| 99 | Phloretin | 44.1 | 273 | MS22→ 167 (100); MS23→ 123 (100); MS24→ 95 (100) | - | P | - | - | - |
| 100 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin(isomer) | 43.0 | 575 | MS22→ 423 (100), 449 (43), 539 (10), 407 (11), 327 (10), 285 (27) 289 (15); MS23→ 285 (100); MS24→ 241 (100), 217 (71), 125 (40) | - | P | - | P | - |
| 101 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin(isomer) | 48.0 | 575 | MS22→ 423 (100), 449 (77), 539 (52), 407 (26), 327 (10), 285 (34) 289 (29); MS23→ 285 (100); MS24→ 241 (100), 125 (53) | - | P | - | P | - |
| 102 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin(isomer) | 49.8 | 575 | MS22→ 447 (56), 423 (50), 449 (30), 539 (61), 407 (23), 327 (18), 285 (33) 289 (27); MS23→ 287 (100), 243 (14); MS24→ 285 (100), 243 (58), 125 (71) | - | P | - | - | - |
| 103 | (Epi)catechin-(4,8′/2,7′)-(epi)catechin(isomer) | 59.9 | 575 | MS22→ 287 (97), 449 (100), 325 (14), 431 (47); MS23→ 285 (96), 125 (100) | - | P | - | P | - |
| 104 | Vanillic acid O-hexoside | 9.6 | 329 | MS22→ 167 (100); MS23→ 152 (100), 123 (54), 108 (23) | - | P | P | P | P |
| 105 | Syringic acid | 11.6 | 197 | MS22→ 182 (100), 153 (18); MS23→ 167 (100); MS24→ 123 (100) | - | P | - | - | - |
| 106 | Malic acid | 3.4 | 133 | MS22→ 115 (100); MS23→ 71 (100) | P | P | P | P | P |
| 107 | Pyruvic acid | 3.8 | 87 | Not fragmented | - | P | - | - | - |
| 108 | Ascorbic acid | 3.9 | 175 | MS22→ 87 (100), 157 (34) | - | P | - | - | - |
| 109 | Cratenacin (isomer) | 41.1 | 619 | MS22 → 413 (100), 293 (59); MS23→ 293 (100); MS24→ 293 (100), 249 (38), 173 (24) | P | - | P | - | - |
| 110 | Cratenacin (isomer) | 42.2 | 619 | MS22→ 413 (100), 293 (51); MS23→ 293 (100); MS24→ 293 (100), 249 (14), 175 (18) | P | - | P | - | - |
| 111 | Cratenacin (isomer) | 49.8 | 619 | MS22→ 413 (100), 293 (50); MS23→ 293 (100); MS24→ 293 (100), 249 (17), 173 (19) | P | P | P | P | - |
| 112 | Vitexin 2″-O-rhamnoside | 37.2 | 577 | MS22→ 413 (100), 293 (41); MS23→ 293 (100); MS24→ 293 (100), 249 (23), 221 (13), 174 (12), 117 (10) | P | P | P | P | P |
| 113 | Isovitexin 2″-O-rhamnoside | 39.9 | 577 | MS22→ 413 (100), 293 (41); MS23→ 293 (100); MS24→ 293 (100), 175 (24) | P | - | - | - | - |
Table 2: HPLC retention times (tR) and MSn fragmentation in negative ion mode of C. laevigata, C. monogyna and the herbal drug phenols (where P=present, F=Fruits and L= Leaves).
In the following section structure assignment of selected compounds is illustrated. Full assignment arguments are provided in the Supporting Information.
Characterization of phenolic acids and phenolic acid glycosides
Eight phenolic acids and seven phenolic acid glycosides were identified in the plant extracts and the herbal drug.
Characterization of phenolic acids: Compounds 1, 44, 106 and 108 with retention times (tR) of 2.9, 32.1, 3.4, 3.9 min and m/z of 191, 163, 133, and 175 were identified as quinic acid, p-coumaric acid, malic acid and ascorbic acid, respectively (Table 1), by comparing their retention times and fragmentation behaviour to the MS/MS spectral data of the corresponding phenolic acids authentic standards. Peaks 92, 98, 105 and 107 (tR 4.2, 7.6, 11.6 and 3.9 min) were assigned as citric acid, salicylic acid, syringic acid and pyruvic acid, respectively. The structural identification of these compounds was based on a comparison of their MS/MS and MSn data (Table 1 and 2) with those reported in literature [26-28]. Phenolic acids have previously been identified and quantified in Crataegus species [29-31].
Characterization of phenolic acids glycosides
Compound 91 (tR 9.0 min) was identified as protocatechuic acid O-hexoside with a pseudomolecular ion [M−H]− of 315.0772 (Table 1). It produced daughter ions at m/z 153.0148 corresponding to protocatechuic acid after the neutral loss of the hexoside group and at m/z 109.0288 by the neutral loss of a hexose moiety followed by the neutral loss of CO2 (44 Da).
Similarly, compounds 93-97 and 104 with tR 11.8, 7.7, 11.3, 20.8 and 9.6 min (Table 2) were assigned as syringic acid O-hexoside, salicylic acid O-galactoside, salicylic acid O-glucoside, homovanillic acid O-hexoside, sinapic acid O-hexoside and vanillic acid O-hexoside, respectively as previously reported [32-36]. Although aromatic acids and phenolic acids have been previously identified and quantified in Crataegus species [29-31], their hexosides derivatives have been reported here for the first time.
Characterization of benzyl alcohol-hexose-pentose
One benzoic acid derivative was detected at tR of 21.6 min and m/z of 401.1455 (C18H25O10) in the plant extracts and regarded as benzyl alcohol-hexose-pentose 23 based on the MS/MS data, which provided a daughter ion at m/z 269.1041 (C13H17O6) resulted from the neutral loss of pentose unit. Moreover, the compound showed a mass error below 1 ppm thus confirming its elemental composition. This compound was already mentioned in the literature [33,37].
Characterization of 3-O-methylellagic acid 4’-(2’’,3’’-di-Oacetyl)- rhamnoside
With retention time 64.8 min, one ellagic acid derivative was detected at m/z 545.0920 (C25H21O14) and was tentatively assign ed as 3-O-methylellagic acid 4’-(2’’,3’’-di-O-acetyl)-rhamnoside 85. Compound 85 produced the MS2 base peak at m/z 315 [M−H−230]− by the neutral loss of a rhamnosyl unit and the two acetyl groups connected and a secondary peak at m/z 300 [M−2H−230−15]− (Table 2) due to the subsequence neutral loss of the methyl group. It produced the MS3 base peak at m/z 300 [ellagic acid−2H]−. Similar ellagic acid derivatives were already mentioned in the literature [38]. To the best of our knowledge, compound 85 was not previously reported in nature.
Characterization of (−)-11-hydroxy-9,10-dihydrojasmonic acid 11-β-D-glucoside
One fatty acid derivative (tR 58.5 min) exhibited a deprotonated molecule at m/z 389.1817 was suggested as (−)-11-hydroxy-9,10- dihydrojasmonic acid 11-β-D-glucoside 82. This compound showed MS2 fragment ion at m/z 227, which indicate the neutral loss of glucose moiety [M−H−162]− (Table 2). It also represented MS/MS fragment at m/z 183.1376 due to the neutral loss of sugar unit followed by the neutral [38] loss of CO2 [M−H−162−44]− (Table 1). The precursor ion has already been reported in the literature in Nicotiana tabacum [39]. To our knowledge this compound has been reported in Crataegus derived herbal medicine for the first time.
Characterization of the trisaccharide di-O-glucosyl-glucitol
Compound 11 (tR 12.1 min), which showed an m/z of 505.1772, was assigned as di-O-glucosyl-glucitol based on the presence of two main fragments at m/z 343, which resulted from a neutral loss of glucosyl molecule [M−H−162]−, and m/z 181 [glucitol−H]− due to the neutral loss of the second glucosyl unit [M−H−324]− (Table 1 and 2). It is worth noting that this compound has been reported in Crataegus for the first time.
Characterisation of flavonoids and flavonoids glycosides
Flavonoids are derived from the shikimate pathway in plant kingdom. They have a basic structure consisting of two aromatic benzene rings separated by an oxygenated heterocyclic ring. Several compounds from different flavonoid classes, such as flavonols, flavanones, flavones and others have been characterized and identified in hawthorn and the herbal drops samples. Crataegus species analyzed in this study showed C-glycosides and O-glycosides flavonoids isomers as represented in Table 1 and 2. All flavonoids O-glycosides showed a neutral loss of the glycan part. In contrast, flavonoid C-glycosides showed a series of characteristic fragments of C-glycosides flavonoids at m/z [M−H−18], [M−H−18]−, [M−H−60]−, [M−H−90]−, [M−H−120]−, [M−H−180]− and [M−H−210]− due to a cross-ring fragmentation of glucosides molecules [40].
Characterization of luteolin derivatives: Compounds 62, 65, 68, 77 and 86 (tR 41.5, 41.9, 45.0, 51.1 and 69.9 min) with m/z of 447.0933, 609.1461, 463.0882, 447.0928 and 301.0349 were assigned as luteolin 7-O-glucoside, quercetin 3-O-(6-O-rhamnosyl-glucoside) (rutin), quercetin 3-O-glucoside, quercetin 3-O-rhamnoside (quercitrin) and quercetin, respectively, after comparison their retention times, UV spectra and MS/MS fragmentation patterns with authentic standards (Table 1).
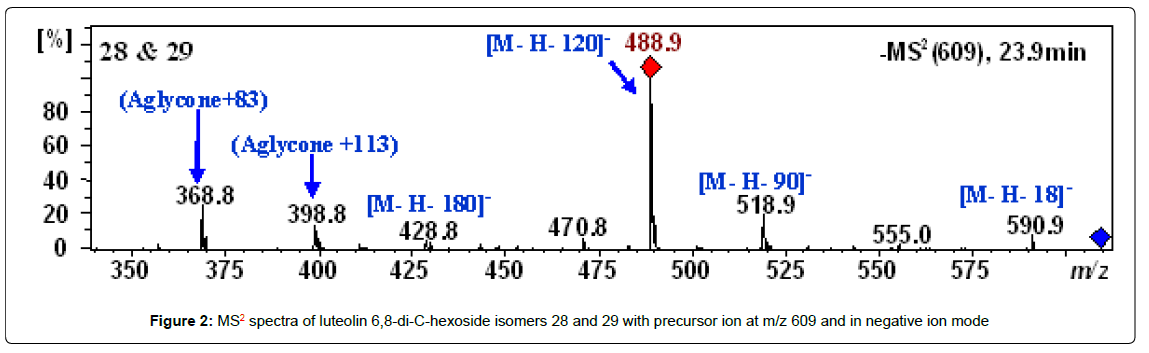
Peaks 28 and 29 with retention times of 23.9 and 30.9 min and m/z of 609.1459 and 609.1462 were tentatively identified as luteolin 6,8-di- C-hexoside isomers. The HPLC-ESI-MS spectra of these peaks showed MS2 fragment ions at [M−H−18]−, [M−H−90]−, [M−H−120]−, [M− H−180]− and [M−H−210]− (Table 2) in addition the fragment ions at m/z 369 (aglycone + 83) and 399 (aglycone + 113), which characterize the luteolin with the residues of the hexoses that remained connected to it (Figure 2) [41].
Compounds 47 and 48, with precursor ions at m/z 447.0929 and 447.0932 and retention times of 33.4 and 34.5 min, respectively, were identified as luteolin C-hexoside isomers (Table 1 and 2). Compound 48 was characterized as luteolin 8-C-glucoside based on the comparison of the retention time and the fragmentation patterns with the authentic standard. From the literature and from our experiments, we have found that the flavonoids glycosylated with galactoside units are less polar than the flavonoids glycosylated with glucoside units [34, 35]. Based on the above argument, the earlier eluted isomer (tR 33.4 min) was assigned as luteolin 8-C-galactoside 47. Luteolin and luteolin glycoside derivatives were already reported in the literature in Crataegus species [31,42].
Characterization of naringenin derivatives: Two naringenin C-hexosides 49 (m/z 433.1136) and 50 (m/z 433.1138) with retention times of 34.1 and 35.2 min were detected in the TOF-MS mode. The MS/ MS product ion scan of these compounds showed characteristic neutral losses of 90 and 120 Da from the parent ion (m/z 433) corresponding to cross-ring cleavages in the sugar unit (Table 1). Although naringenin and naringenin 5,7-diglucoside were already mentioned in the literature in Crataegus species [43,44] these compounds have been reported here for the first time.
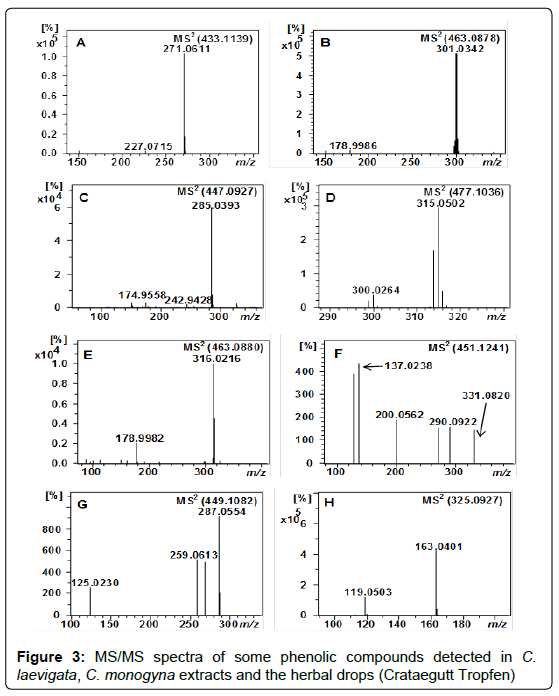
Peak 73 (m/z 433.1139) with a retention time of 46.5 min and molecular formula (C21H22O10) was suggested as naringenin 7-O-glucoside (Table 1 and Figure 3A) based on the presence of fragment ion at m/z 271.0611 [naringenin−H]− and the neutral loss of hexose moiety, resulting in an ion at [M−H−162]− [45]. As far as we know this compound has been reported in Crataegus for the first time.
Characterization of apigenin derivatives: Apigenin 8-C-glucoside (vitexin) 52 and apigenin 6-C-glucoside (isovitexin) 53, vitexin 2″-O-rhamnoside 112, isovitexin 2″-O-rhamnoside 113 and cratenacin (4″′-acetylvitexin-2″-O-rhamnoside) isomers 109-111 were assigned on the bases of the product ion spectrum (Table 1) and comparison with previously reported data [15,45]. Vitexin derivatives are wellknown compounds in Crataegus species [15,46].
Peaks 37, 38, 39 and 40 (tR 28.7, 30.6, 32.2, 35.0 min) exhibited similar molecular formula (C26H28O14). They showed MS/MS fragmentation characteristic to flavone di-C-glycosides with [M− H−18]−, [M−H−60]−, [M−H−90]−, [M−H−120]−, [M−H−180]− and [M−H−210]. The higher intensity of ion at m/z 473 [M−H−90]− relative to the ion at m/z 443 [M−H−120]− indicating 6-C-pentosyl-8-C-hexosyl linkage (Table 2) [47]. Thus, peaks 37, 38 and 40 were characterized as apigenin 6-C-pentosyl-8-C-hexoside isomers. In contrast, peak 39 was tentatively regarded as apigenin 6-C-hexosyl-8-C-pentoside hence it showed the highest intensity (100%) for the ion at m/z 443 [M− H−120]−. From the leaves of C. monogyna and C. pentagyna apigenin di-C-glycosides were isolated previously [48,49].
Peaks 34 and 35 with m/z 593.1507 (C27H30O15) were detected at retention times 27.1 and 36.0 min, respectively (Table 2). These compounds have shown the C-glycoside flavonoids fragmentation patterns similar to the apigenin derivatives mentioned above. Thus, these peaks were proposed as apigenin 6,8-di-C-hexoside isomers. The later eluted isomer 35 was identified as apigenin 6,8-di-C-glucoside (vicenin-3). This compound has been already reported in Crataegus species [46,48,49]. Tentatively, we assigned the earlier eluted isomer (34) as apigenin 6,8-di-C-galactoside (vicenin-4) [34,35]. As far as we know, vicenin-4 has not been reported previously in nature.
In addition to the apigenin C-glycosides, we also identified two apigenin O-glycoside derivatives, namely, apigenin 7-O-rutinoside 74 and apigenin 7-O-glucoside 76 (Table 1). Compound 74 (tR 47.0 min) showed a precursor ion at m/z 577.1557. The neutral loss of rutinoside [M−H−308]− and the appearance of the ion at m/z 269.0466 in the MS/ MS spectra indicated the aglycone apigenin. The acceptable data of MS with the daughter fragment ions of MS/MS have supported compound 74 be apigenin 7-O-rutinoside. This compound was already mentioned in the literature [50]. On another hand, compound 76 (tR 49.1 min) was identified by comparing its retention time and characteristic MS spectral data with apigenin 7-O-glucoside authentic standard.
Characterization of eriodictyol di-C-hexoside: Peak 45 (tR 32.4 min), showed fragment ions of a di-C-hexoside flavonoid (Table 2). These fragments, together with the precursor ion at m/z 611.1615 and the molecular formula C27H31O16 pointed to an eriodictyol di-Chexoside. Eriodictyol has been previously isolated from C. microphylla [44]. Compound 45 was present in all investigated samples.
Characterization of quercetin derivatives: Compounds 24, 51, 67, 75, 79 and 80 with retention times of 21.9, 36.5, 44.0, 47.6, 52.2, 55.7 min and m/z of 625.1411, 755.2037, 463.0878, 433.0772, 505.0984 and 505.0982 were assigned as quercetin 3,7-di-O-hexoside, quercetin 3-O-(2,6-di-O-rhamnosyl-glucoside), quercetin 3-O-galactoside (Figure 3B), quercetin 3-O-xyloside and quercetin O-acetyl hexoside isomers, respectively (Tables 1 and 2) based on the previous studies [25,35,51,52].
Characterization of kaempferol derivatives: Kaempferol derivatives were also identified in C. laevigata, C. monogyna and the herbal drug. The detected ions at m/z 447.0930 and 447.0927, which correspond to peaks 63 and 64, showed a neutral loss of hexosyl moiety resulting in fragments at m/z 285.0393 and 284.0323 (Table 1), corresponded to the aglycone kaempferol. Thus, they were suggested as kaempferol-O-hexoside isomers. Peak 63 (tR 48.3 min) was as assigned as kaempferol 7-O-glucoside after the comparison with the retention time and fragmentation pattern of kaempferol 7-O-glucoside authentic standard. Peak 64 (tR 50.5 min) has been proposed as kaempferol 3-O-glucoside (Figure 3C).
One peak was readily detected with tR of 40.9 min and of m/z 461.0725 in the EIC and was tentatively assigned as kaempferol 3-O-glucuronide 61. It produced the MS2 base peak at m/z 285 [kaempferol−H]− by the neutral loss of a glucuronide residue (176 Da) and the characteristic MS fragments of kaempferol in MS3 (Table 2). This compound has been recently reported by De Rosso et al. in hybrid grapes [53]. It is worth mentioning that this compound has been detected in Crataegus for the first time.
Compound 83 exhibited a deprotonated molecule at m/z 431.0980 (C21H19O10) and MS2 fragment ions at m/z 284 [kaempferol−2H]− (base peak) and m/z 285 [kaempferol−H]− due to the neutral loss of a rhamnoside moiety [M–H–146]–. Based on our previous finding [25] and the acceptable MS data (Table 1 and 2) this compound has been assigned to kaempferol 3-O-rhamnoside.
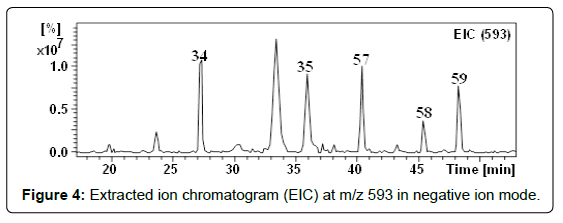
With m/z 593.1512 (C27H29O15), three compounds (57-59) were detected at retention times 40.3, 45.3 and 48.3 min (Figure 4). These compounds showed the same fragmentation patterns. In their MSn they showed ions corresponding to the aglycone kaempferol (Table 2) [54] by the neutral loss of (rhamnosyl-hexoside) [M–H–308]–. Therefore they were suggested to be kaempferol -O-(6-O-rhamnosyl hexoside) isomers. We have found that flavonol-7-O-glycosides elute first followed by flavonol-3-O-glycosides [25]. Based on the order of elution and the similarity of the fragmentation behaviors, isomers 57 and 59 were assigned as kaempferol 7-O-(6-O-rhamnosyl-glucoside) and kaempferol 3-O-(6-O-rhamnosyl-glucoside), respectively. For further evidence, the UV spectrum, the MS fragmentation and the retention time of compound 59 were compared with the kaempferol 3-O-(6-O-rhamnosyl-glucoside) authentic standard. This compound has been previously recorded in quince (Cydonia oblonga) fruits [21]. Considering the elution order in the RF-C18-A column, isomer 58 was regarded as a glycosylated isomer of compound 59 and hence assigned as kaempferol 3-O-(6-O-rhamnosyl-galactoside) [34,35]. Recently, we have detected this compound in I. coccinea [25]. Although kaempferol glycosides derivatives were already mentioned in the literature in Crataegus species [31,55], these compounds have been reported here for the first time.
Characterization of isorhamnetin derivatives: Following the same strategies discussed above, peaks 54, 55 and 56 (tR 40.2, 47.1 and 48.6 min) were assigned as isorhamnetin 7-O-(6-O-rhamnosylglucoside), isorhamnetin 3-O-(6-O-rhamnosyl-galactoside) and isorhamnetin 3-O-(6-O-rhamnosyl-glucoside), respectively (Table 1 and 2). Although an isorhamnetin glycoside derivative has been previously reported by Rodrigues et al. in C. monogyna [56], these compounds have been reported here for the first time.
Peak 72 (tR 45.9 min) with m/z of 477.1036 (C22H21O16), was identified as isorhamnetin O-hexoside according to the MS, MS/MS data (Table 1) and data from the literature [57]. The presence of the fragment ion at m/z 315.0502 (C16H11O7) indicates the neutral loss of a hexosyl moiety and the presence of the aglycone isorhamnetin (Figure 3D).
Characterization of diosmin: Peak 60 (tR 40.6 min) at m/z 577 was tentatively identified as diosmetin 7-O-rutinoside (diosmin), illustrated by the neutral loss of rutinoside unit [M–H –308]– and the presence of fragment ions at m/z 299 (base peak) (Table 2), which can be referred to [diosmetin−H]− [58]. This fragmentation is consistent with that reported by Lin L [59], who also found many isomers of O-glycosylated diosmetin in flower extract of chrysanthemum (Chrysanthemum morifolium Ramat).
Characterization of myricitrin: Compound 66 with retention time of 42.2 min and m/z of 463.0880 was suggested to myricetin 3-O-rhamnoside (myricitrin), relying on the clear neutral loss of rhamnoside moiety (146 Da) and the characteristic fragment ions at m/z 317, 316, 271, 179 and 151, which represented myricetin (Figure 3E, Table 1 and 2) [60]. In addition, the aglycone myricetin was already reported in a recent study in C. cuneata [61]. To our knowledge this compound has been reported in hawthorn for the first time.
Characterization of pectolinarin: The compound at the retention time 51.8 min and a pseudomolecular ion [M–H]–` of 621.1825 was tentatively assigned as pectolinarin (78) based on the acceptable data of MS (Table 1) and the information obtained from the literature [62].
Characterisation of phloretin derivatives
The analysis in TOF-MS mode also showed the presence of phloretin derivatives in Crataegus species and the herbal drug (Crataegutt Tropfen). Compound 81 (tR 53.9 min; m/z 435.1297; C21H24O10) was positively identified as phloretin 2’-O-glucoside (phlorizin) based on the comparison of the retention time and the fragmentation patterns with the authentic standard (Table 1).
Compound 99 (C15H14O5) with retention time of 41.1 min and pseudomolecular ion [M–H]– of m/z 273.0768 was assigned to phloretin relying on the acceptable data of MS (Table 1), fragmentation pattern of MS2/MS3 (Table 2) and the data obtained from literature [63]. As far as we know, phlorizin and phloretin have never been reported in Crataegus species before.
Characterisation of anthocyanidins, proanthocyanidins and their derivatives
Anthocyanidins, proanthocyanidins and their derivatives are wellknown phytoconstituents in Crataegus species. Our findings boosted and were in line with previous studies [15,64,65].
Characterization of epicatechin derivatives: Peak 30 (tR 25.8 min) at m/z 289.0716 was identified as epicatechin by comparison of UV spectrum and retention time with a commercial standard.
Compound 20 at m/z 451.1241 (tR 18.3 min) with molecular formula C21H23O11 was considered as (epi)catechin C-hexoside relying on its MS and MS/MS fragmentation pattern (Figure 3F) and the data obtained from literature [66].
Characterization of B-type proanthocyanidins and their derivatives: B-type proanthocyanidin oligomers were also detected in the herbal medicine and Crataegus samples. They showed UV spectra with λmax 280 nm, characteristic of proanthocyanidins. Thus, peaks 25-27 (C30H26O12), eluted between 22 and 42 min, showed a pseudomolecular ions [M–H]– at m/z 577.1348, 577.1347 and 577.1350 corresponding to dimeric B-type proanthocyanidins [21].
Peaks 42 (tR 26.5 min) and 43 (tR 31.2 min) were detected at m/z 865.1947 and 865.1976 in the EIC. These compounds were assigned as trimeric B-type proanthocyanidin with (epi)catechin monomeric units [21].
On the other hand, compound 46 (tR 32.7 min) had pseudomolecular ion [M–H]– at m/z 561.1402 and an MS/MS fragment ion at m/z 289.0715 (epicatechin), which corresponds to the neutral loss of (epi) afzelechin (272 Da) (Table 1). Thus, this dimer was suggested to (epi) afzelechin-(epi)catechin. This compound has been previously reported in literature and its fragmentation pathway was described [67]. It is worth noting that peak 46 has been reported in Crataegus for the first time.
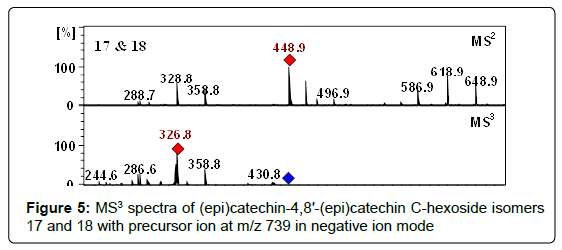
At m/z 739.1880 two compounds (17, 18) were detected at retention times of 17.3 and 21.8 min and were proposed as proanthocyanidin dimers having hexose groups attached to them, showed by the acceptable data of MS (Table 1), fragmentation pattern of MS2/MS3 (Table 2) and the data obtained from literature [68]. Moreover, these peaks represented the characteristic flavan-3-ols-C-glycosides ions at m/z 649 [M−H−90]−, 619 [M−H−120]−, and 589 [M−H−150]− in their MSn (Figure 5). Consequently, compounds 17 and 18 were assigned as (epi)catechin-4,8′-(epi)catechin C-hexoside isomers. Nevertheless, assignment of positions and identity of the hexose units was not possible. According to our knowledge, these isomers have never been reported in Crataegus species before.
Characterization of A-type proanthocyanidins: With m/z 575 (C30H23O12) four peaks were detected at retention times of 48.0, 49.8 and 59.9 min and assigned as dimeric A-type proanthocyanidin, tentatively (epi)catechin-(4,8′/2,7′)-(epi)catechin isomers (100-103) (Table 1 and 2).
We have calculated identical molecular formula (C45H36O18) for peaks 87-90 at m/z 863 (Table 1). They eluted between 30 and 40 min in the developed reverse phase chromatographic method. These compounds were tentatively assigned as trimeric A-type proanthocyanidins with (epi)catechin monomeric units. The suggested structures and fragmentation pathways of A-type proanthocyanidin oligomers have been discussed in previous studies [21,23,25,67].
Characterization of cyanidin derivatives: The detected precursor ion at m/z 287.0561 (C21H21O11+), which correspond to peaks 84 (tR 62.6 min) was assigned as cyanidin based on the acceptable data of MS with the daughter fragment ions of MS/MS (Table 1) in addition to the information previously reported in the literature [69].
With m/z 449.1082 three peaks with identical molecular formula C21H21O11 + were detected at retention times of 27.0, 38.5 and 56.8 min. These compounds showed a neutral loss of hexose moiety resulting in a fragment ion at m/z 287.0554, which corresponds to cyanidin (Table 1). Therefore, they were suggested as cyanidin O-hexoside isomers (31-33). Based on the order of elution these peaks were tentatively identified as cyanidin 7-O-glucoside 31, cyanidin 3-O-galactoside 32 and cyanidin 3-O-glucoside 33 [25,34,35,70]. Although compounds 32 and 33 were already mentioned in the literature in hawthorn [70], compound 31 has been reported here for the first time (Figure 3G).
Characterization of cinchonains Ia: The analysis in TOF-MS mode also showed the presence of flavalignan isomers (21 and 22) in the analyzed samples. These isomers (tR 65.2 and 66.7 min) showed the same fragmentation patterns (Table 1). The acceptable data of MS with the produced fragment ions of MS/MS have directed to suggest the parent ions at m/z 451.1030 and 451.1035 to cinchonains Ia isomers. These data are in accordance with previous data cited in literature [71]. Cinchonains and their higher oligomeric have already been detected in Crataegus species [72].
Characterisation of acylated quinic acid derivatives
Several mono and di-acylated quinic acid derivatives including chlorogenic acids and chlorogenic acids glycosides were also detected in fruits and leaves of C. monogyna and C. laevigata and the traditionally derived drops (Crataegutt Tropfen).
Characterization of chlorogenic acids: Peaks 69 (tR 45.6 min), 70 (tR 47.0 min), and 71 (tR 51.0 min) with identical molecular formula (C25H24O12) were identified as 3,4-di-O-caffeoylquinic acid, 3,5-di- O-caffeoylquinic acid and 4,5-di-O-caffeoylquinic acid (Table 1) by comparison with their commercial standards. 3,5-di-O-caffeoylquinic acid has been previously reported in C. monogyna [56], while 3,4-di- O-caffeoylquinic acid and 4,5-di-O-caffeoylquinic acid have been reported here for the first time.
Four caffeoylquinic acids (2-5), four p-coumaroylquinic acids (12- 16) and one feruloylquinic acid were identified in TOF-MS mode using accurate mass measurements and MS/MS fragmentations (Table 1) in addition to identification keys previously cited as 3-O-caffeoylquinic acid 2, cis-3-O-caffeoylquinic acid 3, 5-O-caffeoylquinic acid 4, cis-5-O-caffeoylquinic acid 5, 3-p-coumaroylquinic acid 12, cis- 3-p-coumaroylquinic acid 13, 4-p-coumaroylquinic acid 14, 5-p-coumaroylquinic acid 15, cis-5-p-coumaroylquinic acid 16 and 5-O-feruloylquinic acid 36 [73-77]. Furthermore, compounds 2 and 4 were confirmed by chromatographic comparisons with authentic standards. Although several chlorogenic acid derivatives were already mentioned in the literature in C. monogyna [43,56], compounds 3, 5, 13 and 15 have been reported here for the first time.
Peak 19 (tR 17.6 min) revealed a deprotonated molecule at m/z 325.0927 and MS/MS fragment ions at m/z 163.0401 and m/z 119.0503 corresponding to the neutral loss of a hexosyl moiety [M−H−162]− followed by the neutral loss of carbon dioxide molecule (CO2). In line with the previous reported MS data [32], this compound was assigned as p-coumaric acid O-hexoside (Figure 3H). Assignment of the position of the hexose (glucose or galactose) was not possible because of the lack of a commercial standard.
At m/z 335.0773 (C16H16O9) one peak was detected at retention time of 29.7 min in the EIC. It generated MS2 fragment ions at m/z 179, 173, 135 and 191 (Table 2). Based on this fragmentation pattern this compound was tentatively assigned as 3-O-caffeoylshikimic acid 41, apart from the ion at m/z 191, interestingly, it showed however similar MS2 and MS3 spectra if compared to the MS2 and MS3 spectra of 3-O-caffeoylshikimic acid previously reported [21,78].
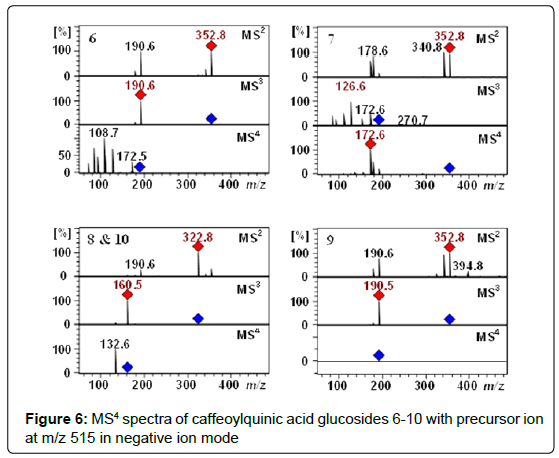
Characterization of chlorogenic acids glycosides: The Q-TOF-MS spectra of peaks 6-9 (tR 11.8, 13.5, 16.5 and 18.1 min) displayed [M−H]− pseudomolecular ion [M–H]– at m/z 515.1406. These compounds with the identical molecular formula C22H27O14 were proposed to be caffeoylquinic acid glucosides (Table 1 and 2). In addition, they showed a typical fragmentation patterns and UV spectra of chlorogenic acids (Figure 6). Thus, these peaks were tentatively proposed as 3-O-(4’-OCaffeoyl glucosyl)quinic acid 6, 4-O-(4’-O-Caffeoyl glucosyl)quinic acid 7, 5-O-(3’-O-Caffeoyl glucosyl)quinic acid 8, 5-O-(4’-O-Caffeoyl glucosyl)quinic acid 9. Recently, we have reported these compounds in Lonicera henryi [79] and Ilex glabra [80].
With retention time of 22.2 min and m/z of 515.1407 (C22H28O14), a further small peak (10) was observed. Compound 10 exhibited fragmentation patterns identical to the 5-O-(3’-O-Caffeoyl glucosyl) quinic acid (8) and we assumed that it might be a cis isomer of compound 8. For confirmation of this isomer, the extract of C. monogyna leaves was irradiated with UV light at 245 nm for 60 min. After irradiation, we found that the cis isomer in the chromatogram as peak with significantly increased intensities if compared to trans isomer from the original plant extract, which confirmed the presence of the cis-5-O-(3’-O-Caffeoyl glucosyl)quinic acid 10 [24]. We have reported the presence of cis derivatives of chlorogenic acids earlier in L. henryi, I. coccinea, quince fruits, coffee leaves, Carlina acaulis, Helianthus tuberosus, Symphyotrichum novae-angliae and Rudbeckia hirta, [21,22,24,25,79]. It is important noting that caffeoylquinic acid glucosides have been reported in hawthorn for the first time.
Conclusion
Crataegus species are medicinal plants used extensively in traditional medicine for the treatment of cardiovascular diseases, arrhythmia and hypertension. Using UPLC-ESI-Q-TOF-MS/MS and HPLC-ESI-MSn, a total of 113 compounds were tentatively identified. To the best of our knowledge 63 of them are described for the first time in Crataegus species and two for the first time in nature. In this context, the obtained results indicate that Crataegus species are rich source of phenolic constituents including simple phenolic acids, chlorogenic acids, proanthocyanidins, flavonoids and flavonoids glycosides.
The current study clearly emphasis the need for re-investigation of medicinal plants using powerful state of the art analytical instrumentation, providing additional information on minor secondary metabolites previously not obtainable.
Acknowledgements
This work was supported by Jacobs University Bremen and Deutscher Akademischer Austausch Dienst (DAAD). We are grateful to Mss Anja Müller for excellent technical support.
References
- Chang WT, Dao J, Shao ZH (2005) Hawthorn: Potential roles in cardiovascular disease. Am J Chin Med 33: 1-10.
- Wang J, Xiong X, Feng B (2013) Effect of Crataegus usage in cardiovascular disease prevention: an evidence-based approach. Evidence-based complementary and alternative medicine :eCAM 149363-149363.
- Tadic VM, Dobric S, Markovic GM, Dordevic SM, Arsic IA, et al. (2008) Anti-inflammatory, gastroprotective, free-radical-scavenging, and antimicrobial activities of hawthorn berries ethanol extract. J Agric Food Chem 56: 7700-7709.
- Blumenthal M, Busse WR, Goldberg A, Gruenwald J, Hall T, et al. (1998) The complete German commission E monographs: Therapeutic guide to herbal medicines. United States of America: Integrative Medicine Communications.
- Halbwirth H (2010) The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway. International Journal of Molecular Sciences 11: 595-621
- Kang NJ, Shin SH, Lee HJ, Lee KW (2011) Polyphenols as small molecular inhibitors of signaling cascades in carcinogenesis. PharmacolTher 130: 310-324.
- Kim D, Lee KW, Lee HJ, Lee CY (2002) Vitamin C equivalent antioxidant capacity (VCEAC) of phenolic phytochemicals. J Agric Food Chem 50: 3713-3717.
- Lavelli V, Hippeli S, Peri C, Elstner EF (1999) Evaluation of radical scavenging activity of fresh and air-dried tomatoes by three model reactions. J Agric Food Chem 47: 3826-3831.
- Mikutis G, Karakoese H, Jaiswal R, LeGresley A, Islam T, et al.(2013) Phenolic promiscuity in the cell nucleus - epigallocatechingallate (EGCG) and theaflavin-3,3 '-digallate from green and black tea bind to model cell nuclear structures including histone proteins, double stranded DNA and telomericquadruplex DNA. Food Funct 4: 328-337.
- Del Rio D, Rodriguez-Mateos A, Spencer JPE, Tognolini M, Borges G, et al (2013) Dietary (Poly)phenolics in Human Health: Structures, Bioavailability, and Evidence of Protective Effects Against Chronic Diseases. Antioxid Redox Signal 18: 1818-1892.
- Pandey KB, Rizvi SI (2009) Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev 2: 270-278.
- Haminiuk CW, Maciel GM, Plata-Oviedo MS, Peralta RM (2012) Phenolic compounds in fruits–an overview. Int J Food Sci Tech 47: 2023-2044.
- Lewandowska U, Szewczyk K, Hrabec E, Janecka A, Gorlach S (2013) Overview of Metabolism and Bioavailability Enhancement of Polyphenols. J Agric Food Chem50: 12183-12199.
- Li Z, Jiang H, Xu C, Gu L (2015) A review: Using nanoparticles to enhance absorption and bioavailability of phenolic phytochemicals. Food Hydrocoll 43: 153-164.
- Belkhir M, Rebai O, Dhaouadi K, Congiu F, Tuberoso CIG, et al. (2013) Comparative analysis of Tunisian wild Crataegusazarolus (yellow azarole) and Crataegusmonogyna (red azarole) leaf, fruit, and traditionally derived syrup: Phenolic profiles and antioxidant and antimicrobial activities of the aqueous-acetone extracts. J Agric Food Chem 61: 12171-12172.
- Cui T, Nakamura K, Tian S, Kayahara H, Tian Y (2006) Polyphenolic content and physiological activities of Chinese hawthorn extracts. Bioscience Biotechnology and Biochemistry 70: 2948-2956.
- Zhang Z1, Chang Q, Zhu M, Huang Y, Ho WK, et al. (2001) Characterization of antioxidants present in hawthorn fruits. J NutrBiochem 12: 144-152.
- Bahorun T, Aumjaud E, Ramphul H, Rycha M, Luximon-Ramma A, et al. (2003) Phenolic constituents and antioxidant capacities of Crataegusmonogyna (Hawthorn) callus extracts. Nahrung 47: 191-198.
- Motilva MJ1, Serra A, Macià A (2013) Analysis of food polyphenols by ultra high-performance liquid chromatography coupled to mass spectrometry: an overview. J Chromatogr A 1292: 66-82.
- Taamalli A1, Iswaldi I, Arráez-Román D, Segura-Carretero A, Fernández-Gutiérrez A, et al. (2014) UPLC-QTOF/MS for a rapid characterisation of phenolic compounds from leaves of Myrtuscommunis L. Phytochem Anal 25: 89-96.
- Karar MGE, Pletzer D, Jaiswal R, Weingart H, Kuhnert N (2014) Identification, characterization, isolation and activity against Escherichia coli of quince (Cydoniaoblonga) fruit polyphenols. Food Res Int 65: 121-129.
- Jaiswal R, Deshpande S, Kuhnert N (2011) Profiling the chlorogenic acids of Rudbeckiahirta, Helianthus tuberosus, Carlinaacaulis and Symphyotrichum novae-angliae leaves by LC-MSn. Phytochem Anal 22: 432-441.
- Jaiswal R1, Jayasinghe L, Kuhnert N (2012) Identification and characterization of proanthocyanidins of 16 members of the Rhododendron genus (Ericaceae) by tandem LC-MS. J Mass Spectrom 47: 502-515.
- Clifford MN, Kirkpatrick J, Kuhnert N, Roozendaal H, Salgado PR (2008) LC-MSn analysis of the cis isomers of chlorogenic acids. Food Chem 106: 379-385.
- Jaiswal R1, Karar MG, Gadir HA, Kuhnert N (2014) Identification and characterisation of phenolics from Ixoracoccinea L. (Rubiaceae) by liquid chromatography multi-stage mass spectrometry. Phytochem Anal 25: 567-576.
- Ratzinger A1, Riediger N, von Tiedemann A, Karlovsky P (2009) Salicylic acid and salicylic acid glucoside in xylem sap of Brassica napus infected with Verticillium longisporum. J Plant Res 122: 571-579.
- de la Luz Cadiz-Gurrea M, Fernandez-Arroyo S, Joven J, Segura-Carretero A (2013) Comprehensive characterization by UHPLC-ESI-Q-TOF-MS from an Eryngiumbourgatii extract and their antioxidant and anti-inflammatory activities. Food Res Int 50: 197-204.
- Rodriguez-Bernaldo de Quiros A, Lage-Yusty MA, Lopez-Hernandez J (2009) HPLC analysis of organic acids using a novel stationary phase. Talanta 78: 643-646.
- Badalica-Petrescu M, Dragan S, Ranga F, Fetea F, Socaciu C (2014) Comparative HPLC-DAD-ESI(+)MS fingerprint and quantification of phenolic and flavonoid composition of aqueous leaf extracts of Cornus mas and Crataegusmonogyna, in relation to their cardiotonic potential. NotulaeBotanicaeHortiAgrobotanici Cluj-Napoca 42: 9-18.
- Tisoncik-Go J, Cordero KS, Rong L (2013) Analysis of oseltamivir resistance substitutions in influenza virus glycoprotein neuraminidase using a lentivirus-based surrogate assay system. Virol Sin 28: 81-91.
- Wu J1, Peng W1, Qin R1, Zhou H2 (2014) Crataeguspinnatifida: chemical constituents, pharmacology, and potential applications. Molecules 19: 1685-1712.
- Abu-Reidah IM, Arraez-Roman D, Quirantes-Pine R, Fernandez-Arroyo S, Segura-Carretero A, et al. (2012) HPLC-ESI-Q-TOF-MS for a comprehensive characterization of bioactive phenolic compounds in cucumber whole fruit extract. Food Res Int 46: 108-117.
- Bystrom LM1, Lewis BA, Brown DL, Rodriguez E, Obendorf RL (2008) Characterization of phenolics by LC-UV/vis, LC-MS/MS and sugars by GC in Melicoccus bijugatus Jacq. 'Montgomery' fruits. Food Chem 111: 1017-1024.
- Kite GC1, Veitch NC (2009) Assigning glucose or galactose as the primary glycosidic sugar in 3-O-mono-, di- and triglycosides of kaempferol using negative ion electrospray and serial mass spectrometry. Rapid Commun Mass Spectrom 23: 3125-3132.
- Schieber A, Keller P, Streker P, Klaiber I, Carle R (2002) Detection of isorhamnetin glycosides in extracts of apples (Malusdomestica cv. "Brettacher") by HPLC-PDA and HPLC-APCI-MS/MS. Phytochem Anal 13: 87-94.
- Vallverdu-Queralt A, Jauregui O, Di Lecce G, Andres-Lacueva C, Lamuela-Raventos RM (2011) Screening of the polyphenol content of tomato-based products through accurate-mass spectrometry (HPLC-ESI-QTOF). Food Chem 129: 877-883.
- Moco S1, Bino RJ, Vorst O, Verhoeven HA, de Groot J, et al. (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205-1218.
- Kim JP1, Lee IK, Yun BS, Chung SH, Shim GS, et al. (2001) Ellagic acid rhamnosides from the stem bark of Eucalyptus globulus. Phytochemistry 57: 587-591.
- Tugizimana F, Steenkamp PA, Piater LA, Dubery IA (2014) Multi-Platform metabolomic analyses of ergosterol-induced dynamic changes in Nicotianatabacum Cells. Plos One 9: e87846.
- March RE, Lewars EG, Stadey CJ, Miao X, Zhao X, et al. (2006) A comparison of flavonoid glycosides by electrospray tandem mass spectrometry. International Journal of Mass Spectrometry 248: 61-85.
- Breiter T, Laue C, Kressel G, Groell S, Engelhardt UH, et al. (2011) Bioavailability and antioxidant potential of rooibos flavonoids in humans following the consumption of different rooibos formulations. Food Chem 128: 338-347.
- Edwards JE1, Brown PN, Talent N, Dickinson TA, Shipley PR (2012) A review of the chemistry of the genus Crataegus. Phytochemistry 79: 5-26.
- Schrall R, Becker H (1977) Production of catechins and oligomericproanthocyanidins in tissue and suspension cultures of Crataegusmonogyna, Crataegusoxyacantha and Ginkgo biloba. Planta Med 32: 297-307.
- Melikoglu G, Bitis L, Mericli AH (2004) Flavonoids of Crataegusmicrophylla. Natural Product Research 18: 211-213.
- Sanchez-Rabaneda F, Jauregui O, Casals I, Andres-Lacueva C, Izquierdo-Pulido M, et al. (2003) Liquid chromatographic/electrospray ionization tandem mass spectrometric study of the phenolic composition of cocoa (Theobroma cacao). Journal of Mass Spectrometry 38: 35-42.
- Nikolov NT (1975) New flavone C biosides from Crataegusmonogyna and Crataeguspentagyna. KhimiyaPrirodnykhSoedinenii 3: 422-423.
- Ernst E (2009) Chinese nutrition therapy: Dietetics in traditional chinese medicine (TCM) edited by JoergKastner. JAMA, J Am Med Assoc 302: 2262-2263.
- Nikolov N1, Seligmann O, Wagner H, Horowitz RM, Gentili B (1982) [New flavonoid-glycosides from Crataegus monogyna and Crataegus pentagyna]. Planta Med 44: 50-53.
- Nikolov N, Dellamonica G, Chopin J (1981) Di-C-glycosylflavones from Crataegusmonogyna. Phytochemistry 20: 2780-2781.
- Hossain MB1, Rai DK, Brunton NP, Martin-Diana AB, Barry-Ryan C (2010) Characterization of phenolic composition in Lamiaceae spices by LC-ESI-MS/MS. J Agric Food Chem 58: 10576-10581.
- Cuyckens F, Ma YL, Pocsfalvi G, Claeys M (2001) Tandem mass spectral strategies for the structural characterization of flavonoid glycosides. Analusis 28: 888-895.
- Ferreres F, Llorach R, Gil-Izquierdo A (2004) Characterization of the inter-glycosidic linkage in di-, tri-, tetra- and penta-glycosylated flavonoids and differentiation of positional isomers by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom 39: 312-321.
- De Rosso M1, Tonidandel L2, Larcher R2, Nicolini G2, DallaVedova A1, et al. (2014) Identification of new flavonols in hybrid grapes by combined liquid chromatography-mass spectrometry approaches. Food Chem 163: 244-251.
- Rodriguez-Perez C, Quirantes-Pine R, Amessis-Ouchemoukh N, Madani K, Segura-Carretero A, et al. (2013) A metabolite-profiling approach allows the identification of new compounds from Pistacialentiscus leaves. J Pharm Biomed Anal 7: 167-174.
- Zhang PC1, Xu SX (2001) [Chemical constituents from the leaves of crataegus pinnatifida Bge. var. major N.E.Br]. Yao Xue Xue Bao 36: 754-757.
- Hevesi Tóth B1, Blazics B, Kéry A (2009) Polyphenol composition and antioxidant capacity of Epilobium species. J Pharm Biomed Anal 49: 26-31.
- Parejo I, Jauregui O, Sanchez-Rabaneda F, Viladomat F, Bastida J, et al. (2004) Separation and characterization of phenolic compounds in fennel (Foeniculumvulgare) using liquid chromatography-negative electrospray ionization tandem mass spectrometry. J Agric Food Chem 52: 3679-3687.
- Hvattum E, Ekeberg D (2003) Study of the collision-induced radical cleavage of flavonoid glycosides using negative electrospray ionization tandem quadrupole mass spectrometry. Journal of Mass Spectrometry 38: 43-49.
- Lin L, Harnly JM (2010) Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat). Food Chem 120: 319-326.
- Toth BH, Blazics B, Kery A (2009) Polyphenol composition and antioxidant capacity of Epilobium species. J Pharm Biomed Anal 49: 26-31.
- Jayakumar JK, Nirmala P, Praveen Kumar BA, Kumar AP (2014) Evaluation of protective effect of myricetin, a bioflavonoid in dimethyl benzanthracene-induced breast cancer in female Wistar rats. South Asian journal of cancer 3: 107-11.
- Schels H, Zinsmeister HD, Pfleger K (1978) Mass-spectrometry of silylated flavonoid glycosides. Phytochemistry 17: 523-526.
- Mena P1, Calani L, Dall'Asta C, Galaverna G, GarcÃa-Viguera C, et al. (2012) Rapid and comprehensive evaluation of (poly)phenolic compounds in pomegranate (Punicagranatum L.) juice by UHPLC-MSn. Molecules 17: 14821-14840.
- Hashempour A, Ghazvini RF, Bakhshi D, Ghasemnezhad M, Sharafti M, et al. (2010) Ascorbic acid, anthocyanins, and phenolics contents and antioxidant activity of ber, azarole, raspberry, and cornelian cherry fruit genotypes growing in Iran. Horticulture Environment and Biotechnology 51: 83-88.
- Svedstrom U, Vuorela H, Kostiainen R, Huovinen K, Laakso I, et al. (2002) High-performance liquid chromatographic determination of oligomericprocyanidins from dimers up to the hexamer in hawthorn. Journal of Chromatography a 968: 53-60.
- Stark T, Hofmann T (2006) Application of a molecular sensory science approach to alkalized cocoa (Theobroma cacao): Structure determination and sensory activity of nonenzymatically C-glycosylated flavan-3-ols. J Agric Food Chem 54: 9510-9521.
- Gu LW, Kelm MA, Hammerstone JF, Beecher G, Holden J, et al. (2003) Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J Agric Food Chem 51: 7513-7521.
- Guimarães R1, Barros L, Dueñas M, Carvalho AM, Queiroz MJ, et al. (2013) Characterisation of phenolic compounds in wild fruits from Northeastern Portugal. Food Chem 141: 3721-3730.
- Song J, Li X, Zeng L, Liu H, Xie M (2011) Determination of cyanidin-3-glucoside (red kernel food colour) in beverages by high performance liquid chromatography and a study of its degradation by quadruple time-of-flight mass spectrometry. Food Additives and Contaminants Part A-Chemistry Analysis Control Exposure & Risk Assessment 28: 1645-1656.
- Trappey A, Bawadi HA, Bansode RR, Losso JN (2005) Anthocyanin profile of mayhaw (Cretaegusopaca). Food Chem 91: 665-671.
- Chen S, Lu C, Zhao R (2014) Qualitative and quantitative Analysis of Rhizomasmilacisglabrae by ultra high performance liquid chromatography coupled with LTQ qrbitrap(XL) hybrid mass spectrometry. Molecules 19: 10427-10439.
- Sendker J, Petereit F, Lautenschlaeger M, Hellenbrand N, Hensel A (2013) Phenylpropanoid-substituted procyanidins and tentatively identified procyanidin glycosides from hawthorn (Crataegus spp.). Planta Med 79: 45-51.
- Jaiswal R1, Kiprotich J, Kuhnert N (2011) Determination of the hydroxycinnamate profile of 12 members of the Asteraceae family. Phytochemistry 72: 781-790.
- Jaiswal R, Kuhnert N (2010) Hierarchical scheme for liquid chromatography/multi-stage spectrometric identification of 3,4,5-triacyl chlorogenic acids in green Robusta coffee beans. Rapid Commun Mass Spectrom 24: 2283-2294.
- Jaiswal R1, Kuhnert N (2011) Identification and characterization of five new classes of chlorogenic acids in burdock (Arctium lappa L.) roots by liquid chromatography/tandem mass spectrometry. Food Funct 2: 63-71.
- Kuhnert N, Jaiswal R, Matei MF, Sovdat T, Deshpande S (2010) How to distinguish between feruloylquinic acids and isoferuloylquinic acids by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 24: 1575-1582.
- Clifford MN1, Johnston KL, Knight S, Kuhnert N (2003) Hierarchical scheme for LC-MSn identification of chlorogenic acids. J Agric Food Chem 51: 2900-2911.
- Jaiswal R1, Sovdat T, Vivan F, Kuhnert N (2010) Profiling and characterization by LC-MSn of the chlorogenic acids and hydroxycinnamoylshikimate esters in maté (Ilex paraguariensis). J Agric Food Chem 58: 5471-5484.
- Jaiswal R, Mueller H, Mueller A, Karar MGE, Kuhnert N (2014) Identification and characterization of chlorogenic acids, chlorogenic acid glycosides and flavonoids from Lonicerahenryi L. (Caprifoliaceae) leaves by LC-MSn. Phytochemistry (Elsevier) 108: 252-263.
- Jaiswal R, Halabi EA, Karar MGE, Kuhnert N (2014) Identification and characterisation of the phenolics of Ilex glabra L. Gray (Aquifoliaceae) leaves by liquid chromatography tandem mass spectrometry. Phytochemistry 106: 141-155.
Relevant Topics
- Analytical Methods
- Biocatalysis and biotransformation
- Bioinorganic Chemistry
- Biological chemistry
- Bioorganic chemistry
- Chemical Biology of Tetracyclines
- Chemotherapeutic Agents
- Enzyme Inhibitor
- Mechanisms of DNA Damage and Repair
- Nanomaterials For Imaging and Drug Delivery
- Natural Product Biosynthesis
- Nucleic Acid Analogs
- Spectroscopic Probes
Recommended Journals
Article Tools
Article Usage
- Total views: 15911
- [From(publication date):
February-2016 - Jul 11, 2025] - Breakdown by view type
- HTML page views : 14409
- PDF downloads : 1502