Review Article Open Access
Update on Methyltransferase Inhibitors of the Dengue Virus and Further Scope in the Field
Wangikar P1, Martis EAF1, Ambre PK1, Nandan S2 and Coutinho EC1*
1Molecular Simulations Group, Department of Pharmaceutical Chemistry, Bombay College of Pharmacy, Kalina, Santacruz (E), Mumbai 400 098, India
2Chemworx, Creative Industries Premises, Kalina, Santacruz [E], Mumbai 400098, India
- *Corresponding Author:
- Evans C Coutinho
Molecular Simulations Group
Department of Pharmaceutical Chemistry
Bombay College of Pharmacy
Kalina, Santacruz (E)
Mumbai 400 098
India
Tel: +91-22-26670905
E-mail: evans@bcp.edu.in
Received date: April 30 , 2015; Accepted date: May 22, 2016; Published date: June 02, 2016
Citation: Wangikar P, Martis EAF, Ambre PK, Nandan S, Coutinho EC (2016) Update on Methyltransferase Inhibitors of the Dengue Virus and Further Scope in the Field. J Emerg Infect Dis 1:108. doi:10.4172/2472-4998.1000108
Copyright: © 2016 Wangikar P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Infectious Disease and Pathology
Abstract
Dengue virus (DENV) is extremely difficult to eradicate and the virus is responsible for a significant disease burden globally wherein several million people are mortally affected. In the quest for developing a treatment of this disease, scientists have described the methyltransferase (MTase) as a potential target. It is responsible for catalysing the methyl transfer from S-adenosyl-L-methionine that results in capping the mRNA at the N7 of a guanine and 2’-O of the first ribonucleotide. However, selectivity is the biggest challenge due to the presence of methyltransferases in humans. This review highlights key breakthroughs made in developing molecules against DENV MTase, and computational strategies that are employed to tackle the selectivity problem. We also present our perspectives on various unexplored and plausible computational techniques that can be employed to enhance tight binding and overcome the selectivity issues.
Keywords
Methyltransferase; E-Pharmacophore modeling; De novo ligand design; S-adenosyl L methionine
Introduction
Every year about 400 million people are infected by the dengue virus (DENV) which causes a significant disease burden to many regions in the world [1]. DENV belongs to the genus Flavivirus. The virus causes dengue fever (DF) and dengue haemorrhagic fever (DHF), both can be lethal [2,3] if left untreated. The virus is transmitted by arthropod vectors, mainly the mosquito Aedes aegypti. There are five distinct serotypes of the dengue virus (DENV1, DENV2, DENV3, DENV4, DENV5) [4,5]. Dengue is endemic in more than 100 countries but as yet there is no clinically approved vaccine or antiviral therapy [6]. In the past very little information was available to develop effective therapy. However, now with advancements in molecular biology, the molecular mechanisms underlying the DENV replication cycle have been unravelled. As a result, many potential protein targets and their functions are now known. One such potential target is DENV methyltransferase, which has been thoroughly validated [7-9].
Role of methyltransferase in DENV
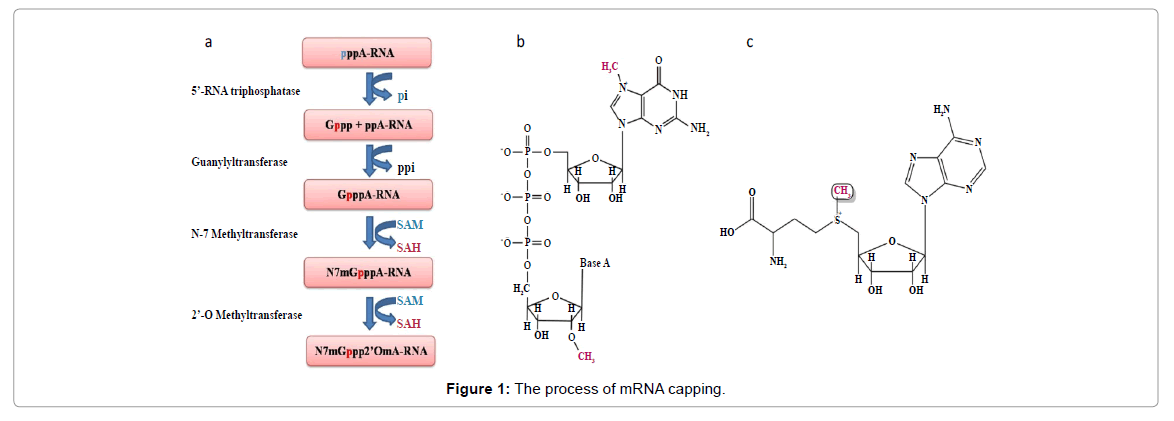
The DENV genome is a positive sense single-stranded RNA, approximately 11 kb long. It comprises of a 5’-untranslated region (UTR), a long open reading frame (ORF), and a 3’-UTR. The ORF encodes a polyprotein that is cleaved by viral and cellular proteases to three structural proteins (capsid, envelope and membrane) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). The non-structural proteins are mainly involved in the replication of the genome and continue the viral progeny. NS3 and NS5 are enzymes, both catalyse important reactions in the life cycle of the dengue virus [7,10,11]. The 5’-end of the dengue mRNA possesses a type 1 cap structure (me7GpppA-me2) that is essential for mRNA stability and effective translation during infection [7,12-14]. NS3 and NS5 have key roles in the formation of the mRNA cap [6]. TheNS5 methyltransferase (MTase) encodes for both N-7 and 2’-O methylation, reactions needed for type 1cap formation (Figure 1a).
The type 1 cap (Figure 1b) is generated via four enzymatic reactions. In the first step, an RNA triphosphatase, hydrolyses the 5’-triphosphate end of the nascent RNA to 5’-diphosphate. In the second step, RNA guanylyl transferase (GTase) caps the 5’-diphosphate RNA with a GMP moiety from GTP. The guanosine cap is then methylated at the N7 position by transfer of a methyl group from S-adenosyl L-methionine (SAM, a cofactor and a methyl donor of MTase; Figure 1c); this is followed by methylation of the 2’-OH position of the first ribose nucleotide (Figure 1b) [2,7].
DENV methyltransferase – new target for drug discovery
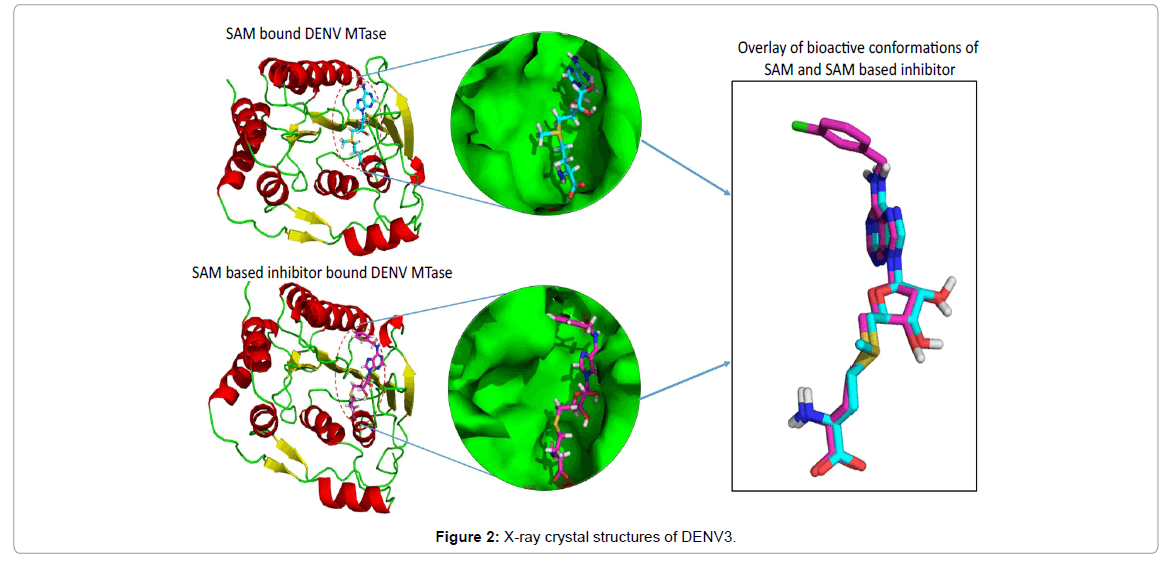
DENV NS5 is the catalytic subunit of the viral replication complex and performs important enzymatic reactions through MTase, as previously mentioned. It is the largest protein (900 amino acids) in DENV with the S-adenosyl-L-methionine (SAM) dependant MTase activity expressed within its N-terminal domain and RNA-dependent RNA polymerase (RdRp) activity within its C-terminal domain [15- 17]. X-ray crystallographic structures reveal that DENV NS5MTase (Figure 2) possesses an overall globular fold, that is divided into an N-terminus region (residues 1–58), a core domain (residues 59–224) and a C-terminus region (residues 225–265) [18]. The core domain comprises of seven β-strands surrounded by four α-helices and two 310 helices. The core domain has two specific binding sites, namely the SAM binding site and the RNA binding site. The N-terminus comprises a helix-turn-helix motif followed by a β-strand and an α-helix, and the C-terminus region consists of an α-helix and two β–strands [16,17,19].
X-ray crystal structures of DENV3 MTase (Figure 2); SAM (magenta), SAM based inhibitor (cyan)) The final two steps of mRNA capping (Figure 1b) are catalysed by DENV MTase using SAM (S-adenosyl- L-methionine) as a methyl donor that generates SAH (S-adenosyl-L-homocysteine) as a by-product [6,14,20]. It has been observed that defects in N7 methylation abolish DENV replication. Therefore, N7 methylation of the viral mRNA cap is considered an essential step in the DENV life-cycle [7,21]. This makes NS5 methyltransferase (MTase) an attractive target for therapeutic interventions [6]. Almost all efforts to find inhibitors against DENV NS5 methyltransferase have been focused on analogs of S-adenosyl-L-methionine.
Strategies used for drug discovery against dengue NS5 MTase
Computational tools serve as powerful approaches to identify high affinity and potent lead molecules. Furthermore, with the availability of high-resolution experimentally solved structures (X-ray or NMR) of proteins/enzymes, the results are far better than using a theoretical model or a lower resolution structure. Such structures help in gaining molecular insights into protein-ligand interactions with atomic level details that enables researchers to accurately predict binding affinities and also to identify new leads by screening and other allied approaches.
Structure based drug design
Using molecular insights of protein structure and function, efficient computational techniques are responsible for the progress made in the drug discovery in current time. Detailed structural information is available for each of the dengue viral proteins. Amongst all, DENV MTase has garnered the maximum attention of researchers in recent years.
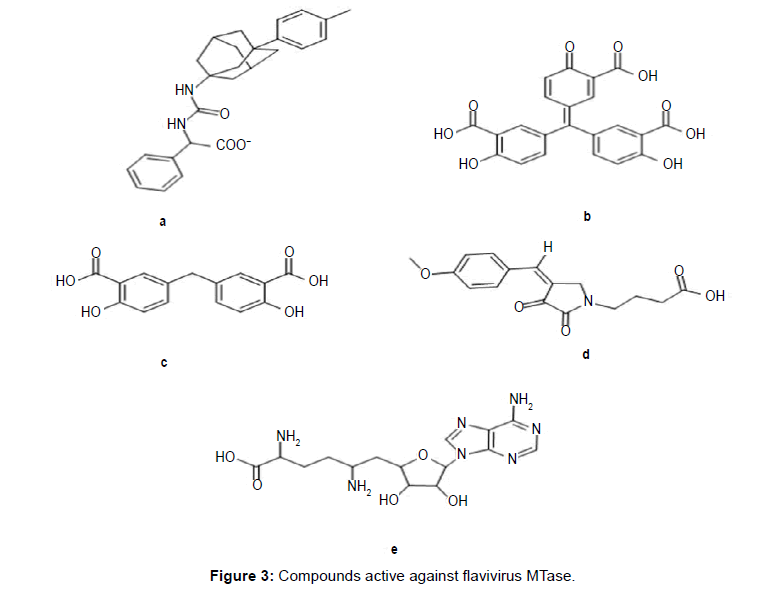
Various attempts have been made in the past to discover effective hits against MTase (Figure 3). Luzhkov et al., reported adamantyl urea derivative (Figure 3a) against MTase (IC50=60 μM) from a database of 2.1 million compounds using high-throughput structurebased screening [22]. Milani et al., by docking simulation found Aurintricarboxylic acid (ATA) as an active molecule against MTase (IC50=2.3 μM) from a Library of Pharmacologically Active Compounds (LOPAC; 1280 commercially available compounds by Sigma–Aldrich) (Figure 3b) [23]. In another approach, Podvinec et al., discovered potent inhibitors of DENV MTase (Figure 3c: IC50=9.5 μM; Figure 3d: IC50=4.4 μM) by screening more than 5 million commercially available compounds (ZINC V5 database and Schrodinger in-house CACDB database) followed by biological assay [16].
Dong et al., reported Sinefungin (Figure 3e) as a competitive inhibitor (IC50=14 μM) of SAM dependent MTase (West Nile Virus) [9]. Lim et al., postulated that Sinefungin inhibits DENV MTase at even lower micromolar concentration (IC50=0.42 μM) [24]. However, selectivity issue and nephrotoxicity associated with this molecule discouraged its marketing as a drug against dengue.
Designing selective inhibitors for flavivirus MTase
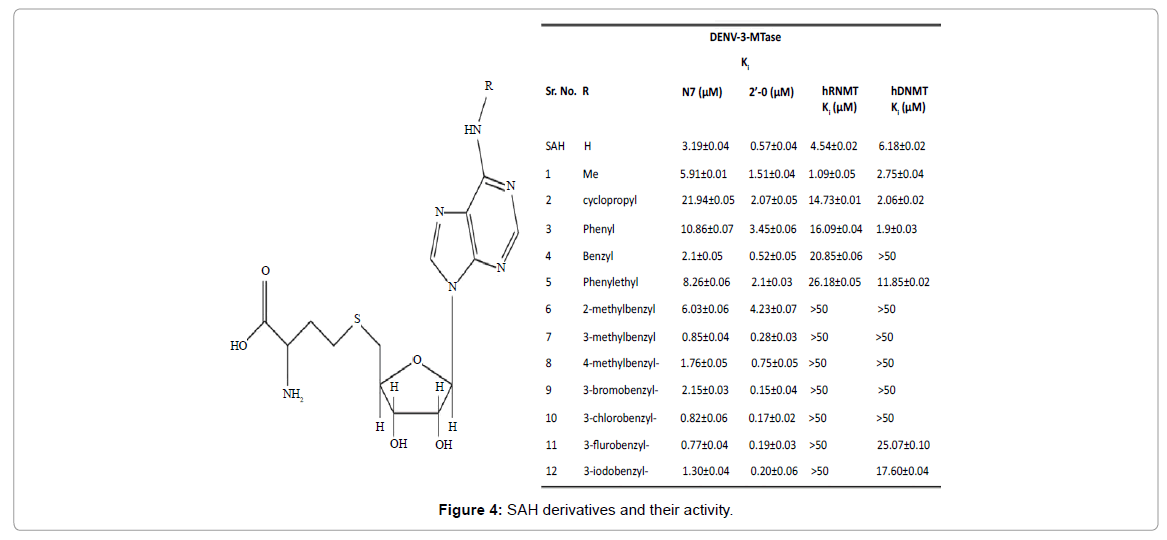
The existence of human MTase and its vital role in normal physiology by methylating proteins as well as DNA and RNA; the design of molecules selective against dengue virus is of prime concern. All MTases use SAM as a cofactor and their core domains are highly conserved. Therefore, any molecule mimicking SAM binding will also inhibit human RNA and DNA MTases, leading to toxicity. Thus to achieve selectivity, considering only the core domain of MTase will be a futile exercise. Hence, incorporating distinct chemical features that could occupy Flavivirus specific pocket [25], in the vicinity of the SAM binding site, which is absent in human MTases will serve as an excellent means to achieve selectivity. Carefully analysing the crystal structures of Flavivirus MTase revealed a conserved cavity located close to the SAM binding site. This cavity largely comprises of hydrophobic residues, namely Phe133, Ile147, Gly148, Glu149, Arg160, Arg163, Val164, and Leu182 (amongst these, Phe133 and Ile147 are part of the SAM binding pocket). This difference can be exploited to introduce selectivity in the MTase inhibitors. Lim et al. confirmed that exploring this cavity could lead to selective inhibition of the Flavivirus MTase, and reported various SAH derivatives (Figure 4) as potent and selective inhibitors of DENV MTase [26].
The N6 substituents at SAH induce conformational changes in the residues (Phe133, Arg163) lining the Flavivirus specific pocket that leads to selective viral MTase inhibition. The benzyl ring at the N6 position (Compound 10) shows improved potency and selectivity for DENV and WNV MTase over SAH. This study showed that hydrophobic groups are well accommodated and are good chemical handles to develop selective inhibitors of DENV MTase.
Similarly, Chen et al. identified nucleoside analogues targeting the WNV MTase selectively [27]. They showed that the selectivity was achieved owing to the substituents occupying the pocket that accommodates methionine part of SAM. Furthermore, they validated their hypothesis by testing their compounds against human MTase, they found that their most active Flavivirus MTase inhibitors (micromolar range), failed to inhibit human MTase even at 300 μM. Both these studies provide good evidence to help researchers design potent and selective inhibitors towards flavivius MTase.
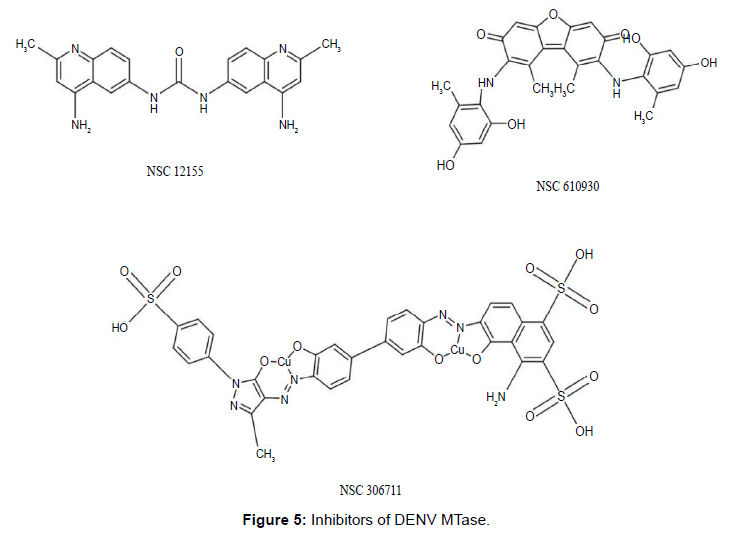
Brecher et al. identified molecules (Figure 5) by screening a diverse set of 1,364 compounds from the National Cancer Institute Developmental Therapeutics Program (NCI DTP) database. NSC 12155 (aminoquinuride; a quinoline derivative) showed good inhibitory activity in a SAM competitive binding assay among six others with good docking scores. The X-ray crystal structure of DENV3 MTase co-crystallised with NSC 12155 (PDB- 5CUQ) has been reported. This X-ray structure reveals that NSC 12155 adapts a curved conformation in the binding site that is dominated by hydrophobic contacts in the pocket [28]. Furthermore, they have also reported inhibitors (NSC 306711 and NSC 610930) identified by virtual screening and docking, additionally confirmed by the biological assay. NSC 306711 was found to be more potent than NSC 610930 in this analysis.
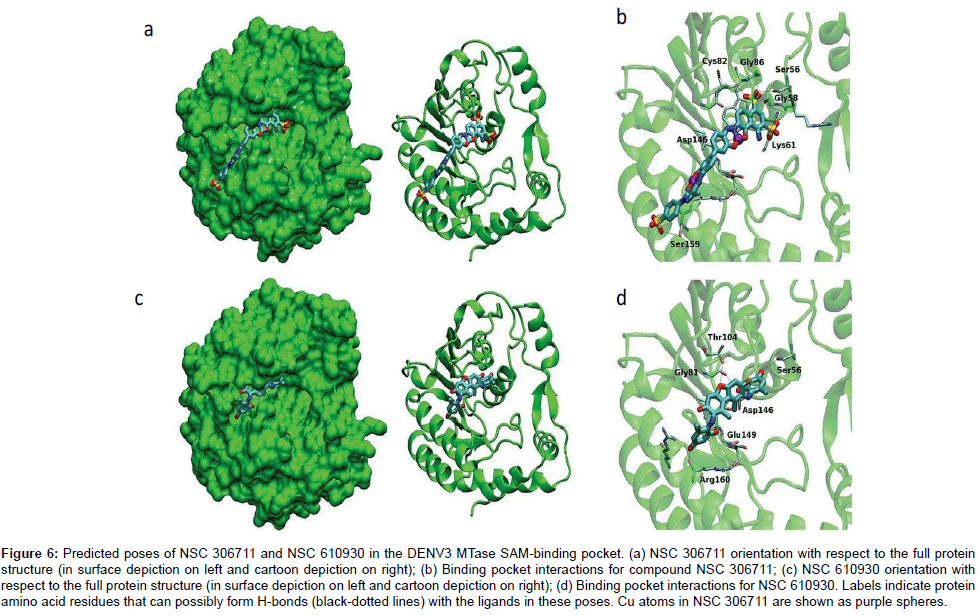
Both NSC306711 and NSC610930 were found to bind in the active site through numerous electrostatic and non-polar contacts when analysed by docking. The larger size of NSC 306711 made possible several close contacts with active site residues compared to the smaller sized NSC 610930. The former makes close contacts with fourteen amino acid residues compared to NSC 610930 that interacts with only nine amino acid residues (Figure 6). The potency of NSC 306711 over NSC 610930 was later confirmed in the biological assay (Table 1) [21].
Figure 6: Predicted poses of NSC 306711 and NSC 610930 in the DENV3 MTase SAM-binding pocket. (a) NSC 306711 orientation with respect to the full protein structure (in surface depiction on left and cartoon depiction on right); (b) Binding pocket interactions for compound NSC 306711; (c) NSC 610930 orientation with respect to the full protein structure (in surface depiction on left and cartoon depiction on right); (d) Binding pocket interactions for NSC 610930. Labels indicate protein amino acid residues that can possibly form H-bonds (black-dotted lines) with the ligands in these poses. Cu atoms in NSC 306711 are shown as purple spheres.
| NSC 12155 | NSC 306711 | NSC 610930 | ||||
|---|---|---|---|---|---|---|
| MTaseenzyme | IC50 N7 (μM) |
IC50 2'-O (μM) |
IC50 N7 (μM) |
IC50 2'-O (μM) |
IC50 N7 (μM) |
IC50 2'-O (μM) |
| DENV2 | 1.2 | 7.6 | 5.3 | 6.9 | 28 | 2.3 |
| DENV3 | 3.4 | 7.7 | 36 | 4.7 | 32 | 21 |
Table 1: Inhibition of DENV MTase by NSC 12155, NSC 306711 and NSC610930.
License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Fragment based drug design
Fragment based approach is yet another useful technique that screens small fragments from various databases, and this can be used to build molecules that are fine tuned to attain better binding affinity as the molecule grows in size [29]. Coutard et al. used such a strategy to screen the Maybridge database of drug-like fragments [30]. They identified 32 fragments confirmed by thermal-shift assay, seven of which were progressed for X-ray crystallographic screening and high concentration screening. Among the seven, five fragments showed promising affinity to DENV MTase (IC50 ranging from 180 μM to 9 mM). Those fragments are six-membered rings, non-fused bicyclic rings, five-membered rings and 5–6 membered fused rings, which likely could be developed into inhibitors with high affinity for MTase [30].
Chemical modification of antiviral molecules
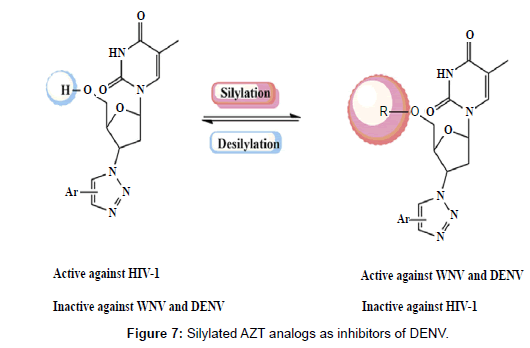
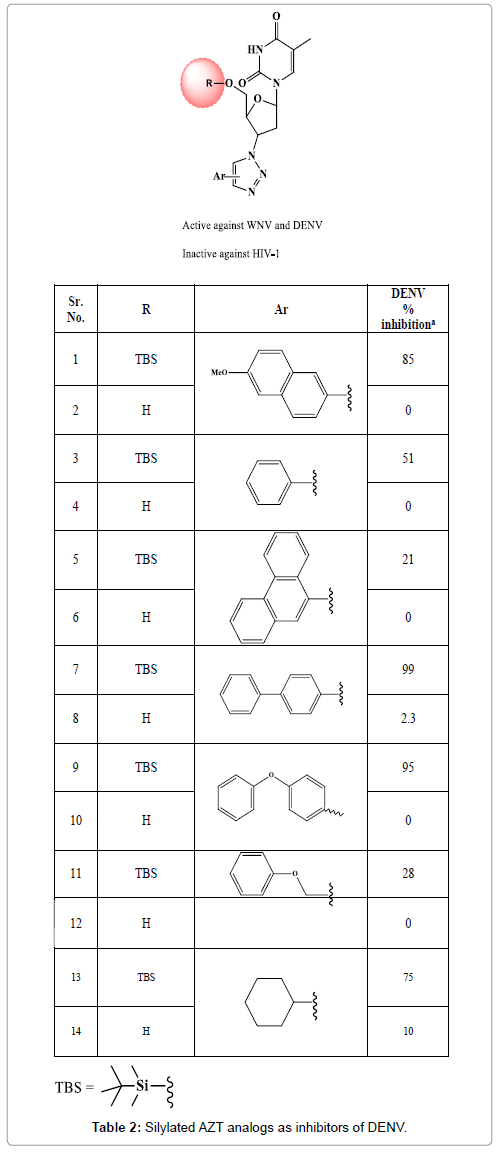
With the aim of achieving selective molecules Vernekar et al., have synthesized 5’ silylated analogs of 3’-azidothymidine-1,2,3-triazole (an inhibitor of HIV-1) (Figure 7). This modification yielded molecules against DENV that reduces the viral titre in a plaque assay (Table 2). The molecules were designed using molecular modelling techniques and following synthesis, the SAM competitive binding assay confirmed that the target of antiviral activity was Flavivirus MTase. The 5’ silyl group and 3’ bulky substituents like biphenyl, diphenylether, p-methoxynaphthyl or cyclohexyl were found to be key features for activity against DENV MTase as revealed by structure activity relationships [31].
Future scope in the field
Till date there is no specific antiviral treatment available for dengue fever and many researchers are striving hard to inch closer to develop a therapeutic agent against dengue. The information extracted from computer simulations or from experiments is considered as a treasure. In recent times many experimental molecules have shown promising results in lowering the viral load. Owing to these breakthroughs, designing inhibitors especially targeting MTase has gained special focus in antiviral drug discovery programs. Besides the strategies that have shown some success in this field in the past, several alternative approaches are yet to be explored for designing novel inhibitors against dengue, the self-limited illness. Some of these alternative approaches are discussed below.
Virtual screening
Virtual screening approaches have proven their prowess in filtering unwanted molecules and identifying potential hits from large databases. This can tremendously reduce the cost of screening inhibitors by saving valuable resources. Databases for virtual screening like ZINC [32,33], Cambridge Structural Database (CSD) [34], PubChem [35], NCI [36] and many more are publicly available free of cost. Moreover, these databases link the molecules to the vendor from where they can be directly purchased in a ready to assay form (DMSO solution).
The caveat in virtual screening is to accurately identify false positives (selection of inactives) and false negatives (rejection of actives). In order to counter this limitation, the DUD database (Database of Useful Decoys) [37] which contains many geometric decoys for various drug targets, enables an assessment of the quality of the screening protocol.
Receptor-based pharmacophore model
The biggest challenge in designing selective inhibitors for DENV MTase is target specificity and hence developing molecules extending to the hydrophobic pocket of DENV MTase is an imperative step to obtain selective molecules (See Structure based drug design) [26]. Receptor based pharmacophore models [38] that combine the feature of active site residues and the residues lining the Flavivirus specific hydrophobic pocket will help to develop selective and potent inhibitors of DENV MTase.
Utilization of pharmacophore modelling tools like Catalyst (Accelrys Inc, USA) or Phase (Schrödinger Inc, USA) help chemists to design selective inhibitors for DENV MTase. Catalyst is one of the most widely used tools to develop a pharmacophore model from a proteinligand complex. It works by generating interaction maps of different features (donor, acceptor, hydrophobes etc.) using LUDI (a program for de novo ligand design). This software also enables one to editor cluster the interactive maps of the target active site using features of receptor so as to retain only essential information. The software easily integrates target structural features with ligand features to produce a model with features crucial for binding [39].
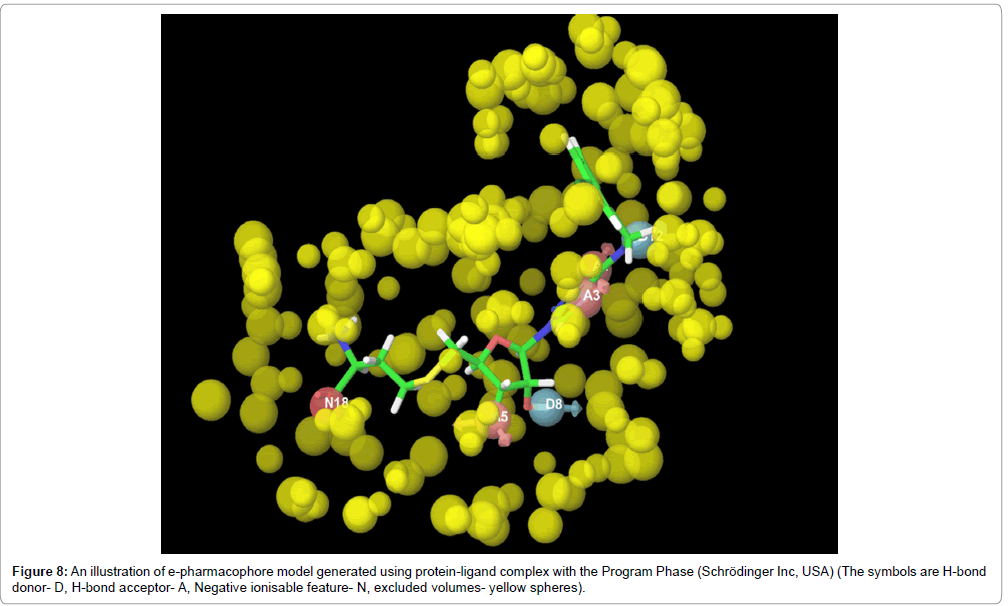
Phase is another software, that derives a pharmacophore model by energy based optimization of a structure based pharmacophore model. The sum of individual Glide XP terms arising from atoms comprises the pharmacophore features and an energetic value is assigned to each feature in Phase. This tool helps to account the induced-fit effects of ligand binding with receptor along with the excluded volume spheres which allows small overlaps with the receptor but avoiding steric clashes (Figure 8) [40].
In recent years, crystal structures of DENV MTase (PDB: 1R6A [41], 3P8Z [26], 5CUQ [28]) with different inhibitors have been published and extensively studied. Using this knowledge, one can generate multiple receptor-based pharmacophore models and combine their features to obtain a single hypothesis. LigandScout is software that provides a platform to combine pharmacophore features to derive a single pharmacophore hypothesis. This unique hypothesis could be utilized to efficiently filter out false positives from various databases [42].
Dynamic pharmacophore model
X-ray crystallographic data depicts a rigid picture of a proteinligand complex and misses out on the dynamic features of the proteinligand interaction. This crystallographic information serves as a good starting point for the development of a dynamic pharmacophore model [43-45]. This approach enables users to incorporate ensembles obtained from MD trajectories to screen potential hits.
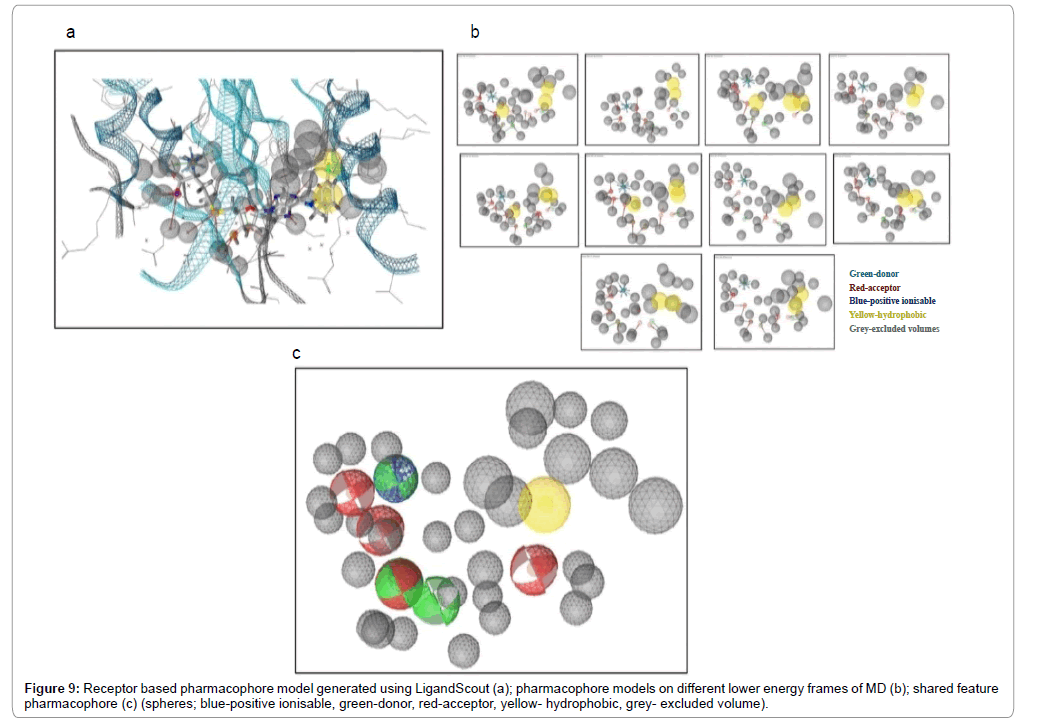
LigandScout enables one to build a dynamic pharmacophore model that can be used to screen numerous databases in search of good hits [42,46]. It extracts ligand information on hybridization state and bond characteristics using an extended heuristic approach along with template based analysis. It derives 3D chemical features from the protein-ligand complex which includes hydrogen bonds donors and acceptors (represented as vectors), negative and positive ionisable groups, aromatic π-π interactions (represented as planes) and lipophilic groups (represented as spheres). Also, excluded volumes are incorporated to count steric clashes and to ensure that lipophilic parts of molecules being screened are matched efficiently (Figure 9). Using LigandScout one can generate multiple pharmacophores using different low energy conformations of the protein-ligand complex derived at different time scales. This combined wealth of information can then be used to derive a ‘shared-feature pharmacophore’.
De novo tools
De Novo drug design is an approximately 20-year old technique that has contributed a lot in drug discovery. It includes specialized programs where a library of small-molecules is built inside the binding pocket by positioning and connecting molecular fragments or by growing a substituent on a starting core [47]. This method can be used on an atom-based or fragment-based approach. The atom-based approach offers a wide range of potential solutions but may yield chemically unstable and improbable suggestions. Fragment-based methods are limited by the database of fragments used, but they are preferred as they produce chemically feasible solutions that are ‘drug like’. Some of the structure based programs are LUDI, LIGBUILD, LEGEND, RECAP, RECORE, BREED, etc. A number of advanced techniques have been established to combine different computational tools with fragment-based approach like CONCERTS (using MD simulation for fragment placing), fragment shuffling (which combines fragments from different ligands in single step), FOG (Fragment Optimized Growth) etc. To assess synthetic feasibility, a scoring function like SYLVIA or CAESA or Route Designer can be used after a de novo predict. In short, de novo techniques may produce better results when assembled with sophisticated adjuvant tools as explained [48,49].
In silico ADMET profile
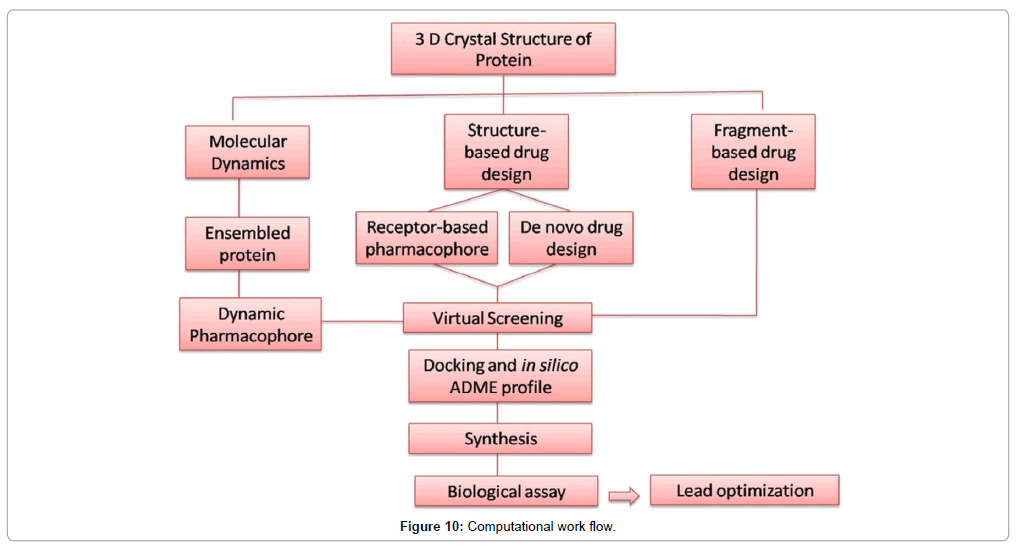
In the case of sinefungin, it was found to be active against DENV MTase in an in vitro assay however, it was found to be nephrotoxicity. As a result, this molecule failed to make it to the clinic. To assess specificity during the drug development cycle of such novel molecules they have to be evaluated for cell cytotoxicity assays. To reduce the laborious cost and time of an experimental procedure, one can verify the ADMET profile of the designed molecules or hits using in silico techniques. Software like QikProp [50,51], TOPKAT [52] can be used for this purpose. The computational work flow is suggested below (Figure 10).
Concluding Remarks
Herein, we have presented some synthetic efforts along with several computational techniques that have been exploited to design potential and selective inhibitors for DENV MTase. The combined approach of pharmacophore modelling and docking analysis could be the method of choice owing to several success stories reported till date. Moreover, receptor-based drug design coalesced with several other computational techniques that assist in predicting drug-like properties, synthetic feasibility as well as ADMET profiles could fructify the search for quality candidates against DENV MTase in the near future.
References
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, et al. (2013) The global distribution and burden of dengue. Nature 496: 504-507.
- Chung KY, Dong H, Chao AT, Shi PY, Lescar J, et al. (2010) Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2'-O methyltransferase activity in dengue virus. Virology 402: 52-60.
- Gubler DJ (1998) Dengue and dengue hemorrhagic fever. ClinMicrobiol Rev 11: 480-496.
- Qi RF, Zhang L, Chi CW (2008) Biological characteristics of dengue virus and potential targets for drug design. ActaBiochimicaetBiophys Sin 40: 91-101.
- Mustafa MS, Rasotgi V, Jain S, Gupta V (2015) Discovery of fifth serotype of dengue virus (DENV-5): A new public health dilemma in dengue control. See comment in PubMed Commons below Med J Armed Forces India 71: 67-70.
- Lim SV, Rahman MB, Tejo BA (2011) Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinformatics 24.
- Dong H, Chang DC, Xie X, Toh YX, Chung KY, et al. (2010) Biochemical and genetic characterization of dengue virus methyltransferase. Virology 405: 568-578.
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, et al. (2007) Structure and function of flavivirus NS5 methyltransferase. J Virol 81: 3891-3903.
- Dong H, Zhang B, Shi PY (2008) Flavivirusmethyltransferase: a novel antiviral target. Antiviral Res 80: 1-10.
- Chambers TJ, Hahn CS, Galler R, Rice CM (1990) Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44: 649-688.
- Klema VJ, Ye M, Hindupur A, Teramoto T, Gottipati K, et al. (2016) Dengue Virus Nonstructural Protein 5 (NS5) Assembles into a Dimer with a Unique Methyltransferase and Polymerase Interface. PLoSPathog 12: 1005451.
- Ghosh A, Lima CD (2010) Enzymology of RNA cap synthesis. Wiley Interdiscip Rev RNA 1: 152-172.
- Noble CG, Li SH, Dong H, Chew SH, Shi PY (2014) Crystal structure of dengue virus methyltransferase without S-adenosyl-L-methionine. Antiviral Res 111: 78-81.
- Decroly E, Ferron F, Lescar J, Canard B (2011) Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10: 51-65.
- Behnam MA, Nitsche C, Boldescu V, Klein CD (2016) The Medicinal Chemistry of Dengue Virus. See J Med Chem.
- Podvinec M, Lim SP, Schmidt T, Scarsi M, Wen D, et al. (2010) Novel inhibitors of dengue virus methyltransferase: discovery by in vitro-driven virtual screening on a desktop computer grid. J Med Chem 53: 1483-1495.
- Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, et al. (2010) Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res 87: 125-148.
- Bussetta C, Choi KH (2012) Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry 51: 5921-5931.
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B (2002) An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21: 2757-2768.
- Shuman S (2000) Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic acid Res MolBiol 66: 1-40.
- Brecher M, Chen H, Liu B, Banavali NK, Jones SA, et al. (2015) Novel Broad Spectrum Inhibitors Targeting the FlavivirusMethyltransferase. PLoS One 10: e0130062.
- Luzhkov VB, Selisko B, Nordqvist A, Peyrane F, Decroly E, et al. (2007) Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2'O)-methyltransferase. Bioorg Med Chem 15: 7795-7802.
- Milani M, Mastrangelo E, Bollati M, Selisko B, Decroly E, et al. (2009) Flaviviralmethyltransferase/RNA interaction: structural basis for enzyme inhibition. Antiviral Res 83: 28-34.
- Lim SP, Wen D, Yap TL, Yan CK, Lescar J, et al. (2008) A scintillation proximity assay for dengue virus NS5 2'-O-methyltransferase-kinetic and inhibition analyses. Antiviral Res 80: 360-369.
- Dong H, Liu L, Zou G, Zhao Y, Li Z, et al. (2010) Structural and functional analyses of a conserved hydrophobic pocket of flavivirusmethyltransferase. J BiolChem 285: 32586-32595.
- Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, et al. (2011) Small molecule inhibitors that selectively block dengue virus methyltransferase. J BiolChem 286: 6233-6240.
- Chen H, Liu L, Jones SA, Banavali N, Kass J, et al. (2013) Selective inhibition of the West Nile virus methyltransferase by nucleoside analogs. Antiviral Res 97: 232-239.
- Brecher M, Chen H, Li Z, Banavali NK, Jones SA, et al. (2015) Identification and Characterization of Novel Broad-Spectrum Inhibitors of the FlavivirusMethyltransferase. ACS Infectious Diseases 1: 340-349.
- Mortier J, Rakers C, Frederick R, Wolber G (2012) Computational tools for in silico fragment-based drug design. Curr Top Med Chem 12: 1935-1943.
- Coutard B, Decroly E, Li C, Sharff A, Lescar J, et al. (2014) Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery. Antiviral Res 106: 61-70.
- Vernekar SK, Qiu L, Zhang J, Kankanala J, Li H, et al. (2015) 5'-Silylated 3'-1,2,3-triazolyl Thymidine Analogues as Inhibitors of West Nile Virus and Dengue Virus. J Med Chem 58: 4016-4028.
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. J ChemInf Model 52: 1757-1768.
- Irwin JJ, Shoichet BK (2005) ZINC-a free database of commercially available compounds for virtual screening. J ChemInf Model 45: 177-182.
- Allen FH (2002) The Cambridge Structural Database: a quarter of a million crystal structures and rising. ActaCrystallogr B 58: 380-388.
- Butkiewicz M, Lowe EW, Mueller R, Mendenhall JL, Weaver CD, et al., (2013) Benchmarking ligand-based virtual high-throughput screening with the PubChem database. Molecules 18: 735-756.
- Islam MA, Pillay TS (2016) Structural requirements for potential HIV-integrase inhibitors identified using pharmacophore-based virtual screening and molecular dynamics studies. MolBioSyst 12: 982-993.
- Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55: 6582-6594.
- Sakkiah S, Thangapandian S, John S, Lee KW (2011) Pharmacophore based virtual screening, molecular docking studies to design potent heat shock protein 90 inhibitors. European journal of Medicinal Chemistry 46: 2937-2947.
- Guner O, Clement O, Kurogi Y (2004) Pharmacophore modeling and three dimensional database searching for drug design using catalyst: recent advances. Curr Med Chem 11: 2991-3005.
- Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, et al. (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20: 647-671.
- Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, et al. (2004) A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate. J BiolChem 279: 35638-35643.
- Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J ChemInf Model 45: 160-169.
- Sohn YS, Park C, Lee Y, Kim S, Thangapandian S, et al. (2013) Multi-conformation dynamic pharmacophore modeling of the peroxisome proliferator-activated receptor γ for the discovery of novel agonists. J Mol Graph Model 46: 1-9.
- Mustata GI, Briggs JM (2002) A structure-based design approach for the identification of novel inhibitors: application to an alanine racemase. J Comput Aided Mol Des 16: 935-953.
- Meagher KL, Carlson HA (2004) Incorporating protein flexibility in structure-based drug discovery: using HIV-1 protease as a test case. J Am ChemSoc 126: 13276-13281.
- Langer T, Wolber G (2004) Virtual combinatorial chemistry and in silico screening: Efficient tools for lead structure discovery? Pure and Applied Chemistry 76: 991-996.
- Schneider G, Fechner U (2005) Computer-based de novo design of drug-like molecules. Nat Rev Drug Discov 4: 649-663.
- Bajorath J (2011) Chemoinformatics and computational chemical biology.
- Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303: 1813-1818.
- Schrodinger (2011) QikProp, New York.
- Ioakimidis L, Thoukydidis L, Mirza A, Naeem S, Reynisson J (2008) Benchmarking the reliability of QikProp. Correlation between experimental and predicted values. QSAR & Combinatorial Science 27: 445-456.
- Cronin MT (2001) Prediction of drug toxicity. 56: 149-151.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 13146
- [From(publication date):
June-2016 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 12069
- PDF downloads : 1077