Research Article Open Access
Universal Primers for Exon-Priming Intron-Crossing (EPIC) PCR on Ribosomal Protein Genes in Marine Animals
Seinen Chow*, Takashi Yanagimoto and Yoji NakamuraNational Research Institute of Fisheries Science, 2-12-4 Fukuura, Kanazawa, Yokohama, Kanagawa 236-8648, Japan
- *Corresponding Author:
- Seinen Chow
National Research Institute of Fisheries Science
2-12-4 Fukuura, Kanazawa
Yokohama, Kanagawa 236-8648, Japan
Tel: +81-45-788-7620
Fax: +81-45-788-5001
E-mail: chow@affrc.go.jp
Received date April 09, 2015; Accepted date May 23, 2015; Published date May 25, 2015
Citation: Chow S, Yanagimoto T, Nakamura Y (2015) Universal Primers for Exon-Priming Intron-Crossing (EPIC) PCR on Ribosomal Protein Genes in Marine Animals. J Marine Sci Res Dev 5:160. doi:10.4172/2155-9910.1000160
Copyright: © 2015 Chow S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
Relatively conserved exon sequences among distant aquatic animal taxa (Copepoda, Echinodermata, and Teleostei) were determined for 12 ribosomal protein genes, and 19 primer pairs were designed for an exon-primed intron-crossing polymerase chain reaction strategy. The universal utility of these primers was evaluated in distant animal species (sea urchin, squid, crab, shark, and tuna). Fragment amplification was confirmed for at least three species for all primer pairs, and single fragment amplification was observed for at least one species in 16 primer pairs. Primer sets presented here may serve as an initial step for isolating single-copy nuclear DNA sequences in a wide variety of marine animals.
Keywords
Universal primers; Ribosomal protein genes; Intron; Single copy nuclear loci; Marine animals
Introduction
Introns may be a good source for finding DNA polymorphisms in the eukaryotic genome as they rapidly accumulate mutations without greatly affecting gene function. Exon-primed intron-crossing (EPIC) polymerase chain reaction (PCR) has been developed and applied to find genetic polymorphisms in a wide variety of animals [1-5]. Primers designed to target conserved areas may have a wide utility across a broad range of animal taxa [6-10]. The universality of the primers for EPIC PCR depends on the conservation of exonic sequences, and the efficiency of PCR amplification may be primarily affected by gene copy number, including pseudogenes. Genes encoding ribosomal proteins (RPs) may be highly suitable for designing universal primers, as RPs are the most highly conserved genes among eukaryotes and are typically encoded by a single gene [11,12]. Indeed, the coding sequences of RP genes are reported to be highly conserved among humans, fruit flies, and nematodes [13]. In this study, we designed 19 primer pairs in exons of 12 RP genes and demonstrated the universal utility of these primer pairs using samples of distantly related aquatic animal species.
Materials and Methods
Eighty different ribosomal protein genes were mapped to human chromosomes [14], and cDNA data for the homologous genes of the copepod (Lepeophtheirus salmonis, Acartia pacifica, or Calanus helgolandicus), sea urchin (Strongylocentrotus purpuratus or Paracentrotus lividus), and fish (Danio rerio or Ictalurus punctatus) were obtained from the Gen Bank database. RP genes that were not found in all three of these reference animal taxa were not analyzed. BLAST searches [15] using these reference cDNA sequences against the draft genome sequences of the Pacific blue fin tuna [Thunnus orientalis; accession nos. BADN01000001-BADN01133062 in the DNA Data Bank of Japan (DDBJ)] [16] detected partial sequences of 31 RP genes among 35 bluefin tuna scaffolds. For each of the 31 genes detected, a multiple sequence alignment was constructed by ClustalX version 1.83 [17], and conserved regions suitable for designing universal primers were surveyed. Crude DNA extractions of the long-spined sea urchin (Diadema savignyi), neon flying squid (Ommastrephes bartramii), golden crab (Chaceon granulatus), mako shark (Isurus oxyrinchus), and skipjack tuna (Katsuwonus pelamis) derived from our laboratory collection were used for evaluating the universal utility of the primers. PCR amplifications were performed in a 12 μL final volume containing 1 μL of template DNA, 1.2 μL 10x buffer, 1 mM of each deoxynucleotide phosphate (dNTP), 0.4 μM of each primer, and 0.5 units of EX Taq polymerase (Takara, Japan). The same reaction conditions were applied for all primer pairs in which the reaction mixtures were preheated to 94°C for 4 min, followed by 35 amplification cycles (94°C for 30 sec, 55°C or 58°C for 30 sec, and 72°C for 50 sec), with a final extension at 72°C for 7 min. PCR products were electrophoresed on a 1.5% agarose gel (UltraPure Agarose, Invitrogen, Carlsbad CA), stained with ethidium bromide and photographed.
Results and Discussion
Relatively conserved exon sequences among distant reference animal taxa were determined for 12 genes in which 19 primer pairs (A–S) were designed for EPIC PCR (Table 1). To design the primers, we adopted a primer length of 20 bases, a moderate GC content (approximately 50%), and usually less than three degenerated bases, and because the conserved regions among the variety of taxa were not extensive, there were limited options for selecting primer positions.
| Pair | Gene | Primer | Orientation | Primer sequence (5'-3') | Distance* |
|---|---|---|---|---|---|
| A | S2 | RPS2exCF | forward | CCHGTNCCCAAGAAGCTGCT | 70 |
| S2 | RPS2exDR | reverse | GGTYTCCTTCCASAGRTCAG | 43 | |
| B | S3 | RPS3ex1F | forward | AAATCTCVAAGAAGAGRAAG | 2 |
| S3 | RPS3ex2R | reverse | ACRCCGGAGTANCCATCCTC | 62 | |
| C | S3 | RPS3ex2F | forward | GAGGATGGNTACTCCGGYGT | 53 |
| S3 | RPS3ex3R | reverse | CKCTTCTGVACMACAGCGGT | 48 | |
| D | S3 | RPS3ex3F | forward | ACCGCTGTKGTBCAGAAGMG | 30 |
| S3 | RPS3ex4R | reverse | GTCGCAACCTTCTCAGCRTA | 24 | |
| E | S3 | RPS3ex4F | forward | TAYGCTGAGAAGGTTGCGAC | 74 |
| S3 | RPS3ex5R | reverse | AASCKCAGRACACCATARCA | 6 | |
| F | S14 | RPS14exAF | forward | GCCTCMTTYAACGAYACCTT | 29 |
| S14 | RPS14exBR | reverse | CKGTCRGCCTTYACCTTCAT | 30 | |
| G | S17 | RPS17exBF | forward | AGWGGMATCTCCATCAARCT | 49 |
| S17 | RPS17exCR | reverse | GGRTCRACCTCGATGATSTC | 18 | |
| H | S23 | RPS23exAF | forward | TCACACGCCAARGGCATYGT | 15 |
| S23 | RPS23exBR | reverse | ATRGCAGAGTTRGGCTGCTT | 14 | |
| I | S23 | RPS23exBF | forward | AAGAACGGCAARAAGATCAC | 42 |
| S23 | RPS23exCR | reverse | TGACCYTTACGWCCRAATCC | 26 | |
| J | S27 | RPS27exAF | forward | GCTAARATYCARGAYAAGGA | 4 |
| S27 | RPS27exBR | reverse | GTGCGKCCRTCYTCCAGCTG | 43 | |
| K | L3 | RPL3exAF | forward | GGCTACAAGGCYGGYATGAC | 39 |
| L3 | RPL3exBR | reverse | CAGTTYTTGTAGAAKCGACG | 149 | |
| L | L3 | RPL3exBF | forward | GCGTCGMTTCTACAARAACT | 2 |
| L3 | RPL3exCR | reverse | GTGAARGCCTTCTTCTYGGA | 9 | |
| M | L3 | RPL3exDF | forward | CAAGGGWCACGGRTDCAAGG | 2 |
| L3 | RPL3exER | reverse | GTCTTRCGGGGMAGCTTCTT | 25 | |
| N | L8 | RPL8exBF | forward | AGTTCATCTACTGCGGCAAG | 37 |
| L8 | RPL8exCR | reverse | ATGAYGGTDCCYTCAGGCAT | 5 | |
| O | L9 | RPL9exCF | forward | RAACTTCYTGGGRGAGAAGT | 110 |
| L9 | RPL9exDR | reverse | ACDGTGGTGGCYTGCTGGAT | 10 | |
| P | L11 | RPL11exAF | forward | CARACGCCHGTCTTCTCCAA | 5 |
| L11 | RPL11exBR | reverse | AAGGATCKYACAGTGTAGCG | 4 | |
| Q | L11 | RPL11exBF | forward | CGCTACACTGTRMGATCCTT | 87 |
| L11 | RPL11exCR | reverse | CCCARRTCGATGTGYTCCTG | 65 | |
| R | L17 | RPL17exAF | forward | GAGWCMGCTCAGGCCATCAA | 132 |
| L17 | RPL17exBR | reverse | TCGGCRCTCTTCTTRGGCCA | 8 | |
| S | L24 | RPL24exBF | forward | AGMCGCAARCACAAGAAGGG | 13 |
| L24 | RPL24exCR | reverse | ACYTCAGGCTTCTGRTTCCT | 85 |
*Number of nucleotides between 3’-end of the primers and start or end of the target introns
Table 1: Sequences of primers for amplifying 19 introns of 12 ribosomal protein genes.
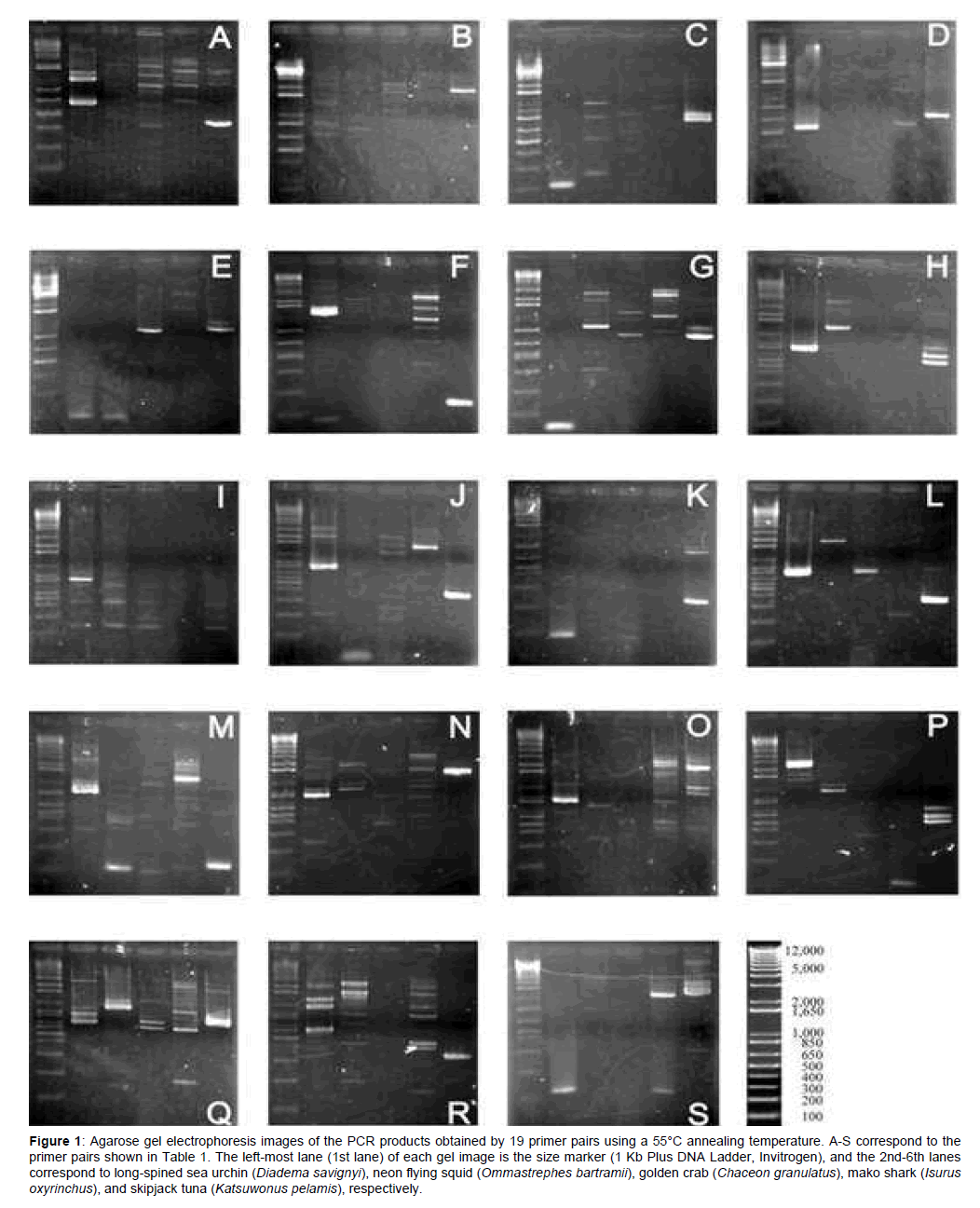
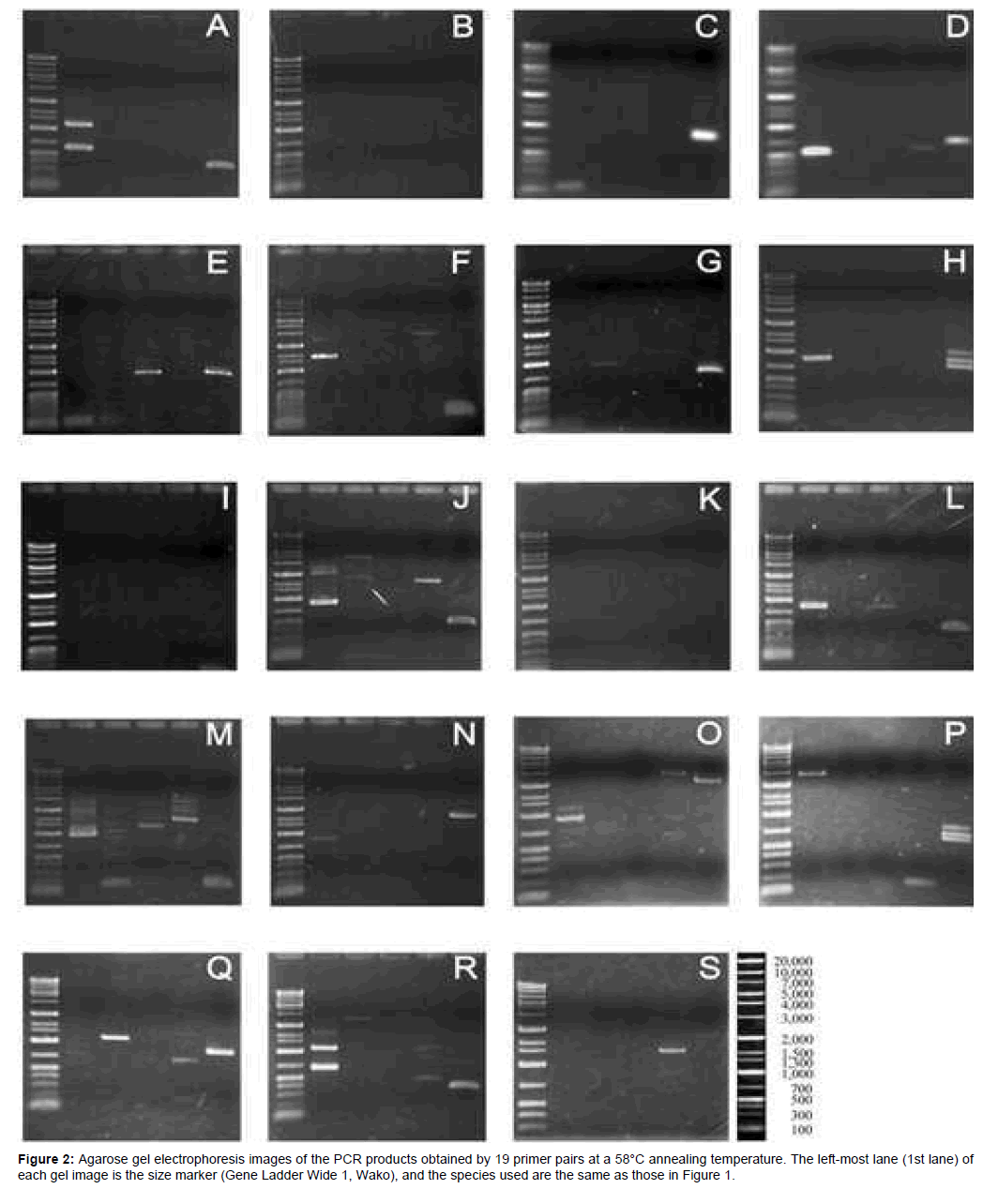
At a lower annealing temperature (55°C), amplification of single to multiple fragments ranging from approximately 100 to 4,000 bp (fragments smaller than 100 bp were not counted) was observed in at least three taxa for each primer pair (Figure 1). The number of amplified fragments decreased at the higher annealing temperature (58°C), to the extent that no amplified fragments were obtained for three primer pairs (B, I, and K; Figure 2). Amplification failure may be attributable to primer mismatch, large numbers of processed pseudogenes [18], and/or non-specific annealing, which may be responsible for multiple fragment amplification. At the higher annealing temperature, the number of primer pairs which amplified a single fragment was 13 pairs in tuna, followed by 8 in sea urchin, 6 in shark, 3 in squid, and only 2 in crab. Higher scores observed in tuna and sea urchin are reasonable, since RP gene data of teleost and sea urchin were used to design primers. The lowest result in crab may indicate distant relationship between copepod and decapod.
Figure 1: Agarose gel electrophoresis images of the PCR products obtained by 19 primer pairs using a 55°C annealing temperature. A-S correspond to the primer pairs shown in Table 1. The left-most lane (1st lane) of each gel image is the size marker (1 Kb Plus DNA Ladder, Invitrogen), and the 2nd-6th lanes correspond to long-spined sea urchin (Diadema savignyi), neon flying squid (Ommastrephes bartramii), golden crab (Chaceon granulatus), mako shark (Isurus oxyrinchus), and skipjack tuna (Katsuwonus pelamis), respectively.
Two to three amplicons (13 in total) from five primer pairs (D, E, G, L, and Q) showing a single fragment amplification at the higher annealing temperature (Figure 2) were selected and subjected to direct nucleotide sequencing using PCR primers. Partial or complete nucleotide sequences were successfully determined for eight (one each from primer pairs D, E, and G; two from L; and three from Q) out of 13 amplicons. Conserved exon sequences and expected exon–intron motifs for the respective RP genes were confirmed in six amplicons from D, E, G, and Q, whereas these were not determined in two amplicons from L due to the short distance between the primer annealing site and an intron (Table 1). All sequences are available in DDBJ and GenBank under the accession numbers LC055480–LC055485, LC055565, LC055566.
Although intensive investigation into the optimal PCR conditions was not performed, amplification failure and multiple fragment amplification may be improved by controlling the annealing temperature or modifying the primer sequences for taxa of interest.
Thus, the present study offers new nuclear primers potentially applicable to a wide variety of marine animal taxa, which may be an initial step toward isolating single-copy nuclear DNA sequences in a wide variety of marine animals. Subsequent polymorphism detection may also provide a more informative database for genetic species identification and population study.
Acknowledgements
This work was partially supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) (No. 25450292) from the Ministry of Education, Science, Sports, and Culture, Japan.
References
- Lessa EP (1992) Rapid survey of DNA sequence variation in natural populations. MolBiolEvol 9: 323-330.
- Palumbi SR, Baker CS (1994) Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. MolBiolEvol 11:426-435.
- Corte-Real HBSM, Dixon DR, Holland PWH (1994) Intron-targeted PCR: a new approach to survey neutral DNA polymorphism in bivalve populations. Mar Biol 120: 407-413.
- Chow S, Clarke S, Nakadate M, Okazaki M (2007) Boundary between the north and south Atlantic populations of the swordfish (Xiphiasgladius) inferred by a single nucleotide polymorphism at calmodulin gene intron. Mar Biol 152: 87-93.
- Daguin C, Bonhomme F, Borsa P (2001) The zone of sympatry and hybridization of Mytilusedulis and M. galloprovincialis, as described by intron length polymorphism at locus mac-1. Heredity 86: 342-354.
- Chow S, Hazama K (1998) Universal PCR primers for S7 ribosomal protein gene introns in fish. MolEcol 7: 1255-1256.
- Chow S, Nakadate M (2004) PCR primers for fish G6PD gene intron and characterization of intron length variation in the albacore Thunnunsalalunga. MolEcol Notes 4: 391-393.
- Hassan M, Lemaire C, Fauvelot C, Bonhomme F (2002) Seventeen new exon-primed intron crossing polymerase chain reaction amplifiable introns in fish. MolEcol Notes 2: 334-340.
- Pinho C, Rocha S, Carvalho BM, Lopes S, Mourão S, et al. (2010) New primers for the amplification and sequencing of nuclear loci in a taxonomically wide set of reptiles and amphibians. Cons Genet Res 2: 181-185.
- Jennings BM, Etter RJ (2011) Exon-primed, intron-crossing (EPIC) loci for five nuclear genes in deep-sea protobranch bivalves: primer design, PCR protocols and locus utility. MolEcol Res 11: 1102-1112.
- Wool IG (1979) The structure and function of eukaryotic ribosomes. Ann Rev Biochem 48:719-54.
- Wool IG, Chan Y-L, Gluck A (1995) Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol 73: 933-947.
- Yoshihama M, Uechi T, Asakawa S, Kawasaki K, Kato S, et al. (2002) The human ribosoma protein genes: sequencing and comparative analysis of 73 genes.Genome Res 12: 379-390.
- Uechi T, Tanaka T, Kenmochi N (2001) A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics 72: 223-230.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J MolBiol 215: 403-410.
- Nakamura Y, Mori K, Saitoh K, Oshima K, Mekuchi M, et al. (2013) Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. PNAS 110: 11061-11066.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882.
- Duvov KP, Perry RP (1984) The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell 37: 457-468.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 15215
- [From(publication date):
August-2015 - Apr 01, 2025] - Breakdown by view type
- HTML page views : 10580
- PDF downloads : 4635